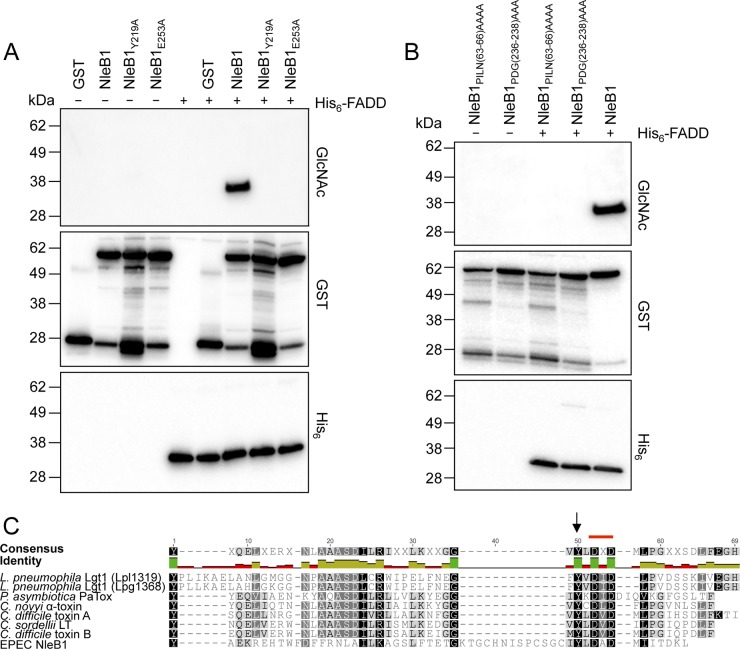
FIG 3.
Loss of enzymatic activity of NleB1 mutants. (A and B) Immunoblots of recombinant protein incubation mixtures from an in vitro assay for NleB1-mediated GlcNAc modification of FADD. Recombinant GST-NleB1 or the mutants and His-FADD were incubated alone or together in the presence of 1 mM UDP-GlcNAc. GlcNAcylation of FADD was tested by immunoblotting with anti-GlcNAc antibodies, and the presence of the GST and His fusion proteins was detected by immunoblotting with anti-GST and anti-His antibodies. The immunoblots are representative of those from at least three independent experiments. (C) Sequence similarity of the NleB1 central region with the catalytic region of glycosyltransferases from Clostridium, Legionella, and Photorhabdus species. A multiple-sequence alignment of the amino acid sequence of the region surrounding the DXD motif (marked with a red line) of EPEC E2348/69 NleB1 (residues 176 to 236; GenBank accession number CAS10779) with different bacterial glycosyltransferases was generated using MUSCLE (28) through the Geneious tool (29). The glycosyltransferases included Legionella pneumophila Lgt1 (Lpl1319 [residues 209 to 260; PDB accession number 2WZG] and Lpg1368 [residues 209 to 260; GenBank accession number Q5ZVS2]), Photorhabdus asymbiotica PaTox (residues 2245 to 2290; GenBank accession number CAQ84322), Clostridium novyi alpha toxin (residues 253 to 296; GenBank accession number Q46149), Clostridium sordellii LT (residues 255 to 298; GenBank accession number Q46342), Clostridium difficile toxin A (residues 253 to 299; GenBank accession number CAC03681), and Clostridium difficile toxin B (residues 255 to 298; GenBank accession number P18177). Black arrow, the Tyr219 residue in EPEC NleB1 conserved in the other glycosyltransferases. The weighted shading of the highlights indicates the percent similarity of the aligned residues, with darker colors indicating the highest conservation. The identity across all sequences for every residue is displayed above the aligned sequences: green, the residue at the given position is the same across all sequences; yellow, less than complete identity; red, very low identity for the given position. Hyphens indicate gaps inserted by Geneious on the basis of the Blosum62 substitution matrix to generate the alignment.