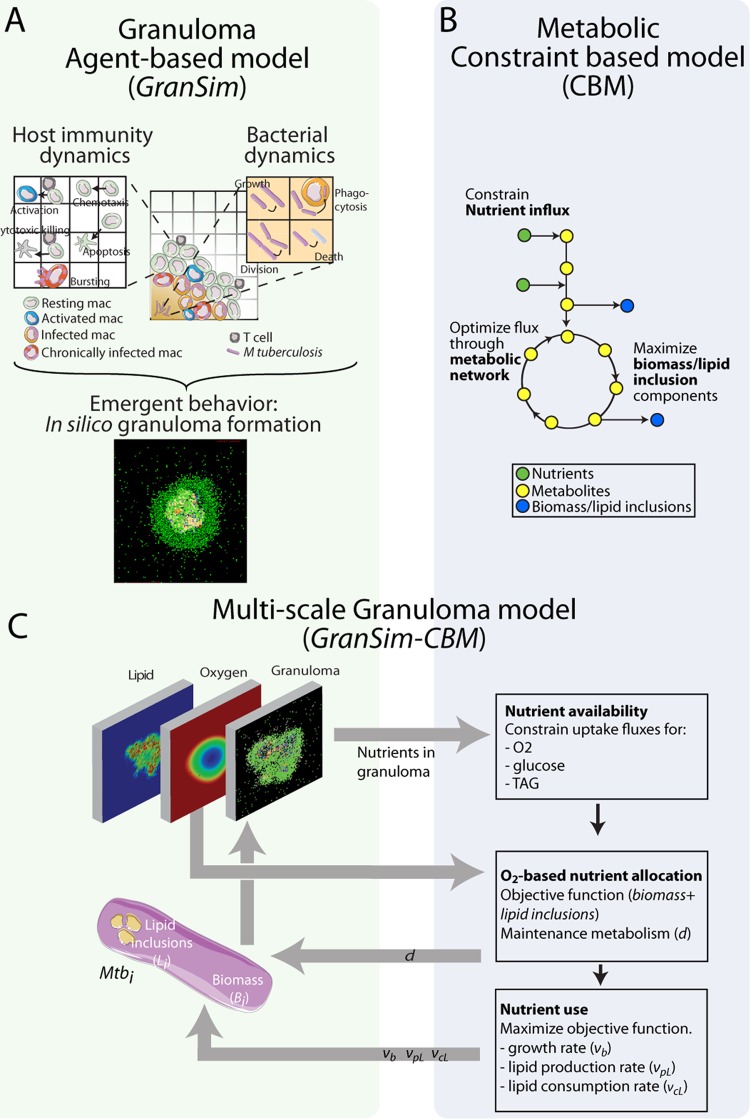
FIG 2.
Multiscale model system bridging metabolic scale to tissue scale. (A) GranSim, our agent-based model of granuloma formation and function, incorporates host immune functions (see references 34–36, and 37 for details) as well as bacterial dynamics for the first time on an individual-bacterium level. In silico granulomas are an emergent behavior of the system. mac, macrophages. (B) The constraint-based model (CBM) uses a stoichiometric matrix representing the metabolic network of M. tuberculosis (45) to predict growth rates based on the bacterial objective function (growth versus lipid inclusion production) (13). (C) The combination model GranSim-CBM tracks granuloma formation and environmental nutrient conditions (oxygen, TAG, and glucose) (left) and uses the CBM to predict growth rates and lipid inclusion formation for each bacterial agent based on its local environment and internal lipid inclusion stores (right). Mtbi, ith bacterial agent.