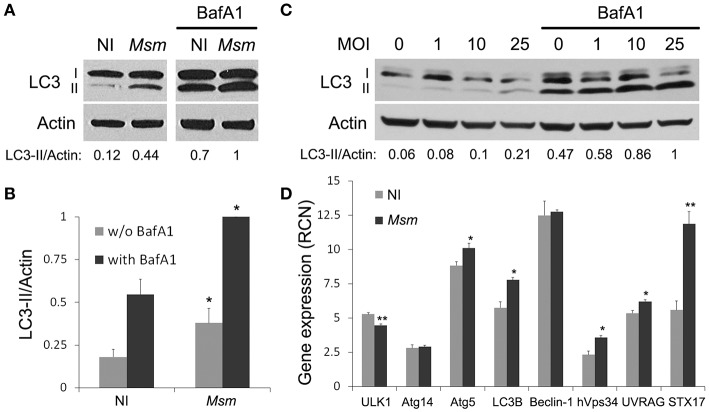
Figure 1.
M. smegmatis induces autophagy in THP-1 macrophages. (A,B) LC3 immunoblotting of THP-1 macrophages infected (Msm) or not (NI) with M. smegmatis. Differentiated THP-1 cells were incubated with M. smegmatis at multiplicity of infection (MOI) 25 for 2 h, washed, and then incubated for 1 h and 30 min with 100 nM Bafilomycin A1 (with BafA1) or DMSO control (w/o BafA1). Cells were lysed and analyzed by immunoblotting with anti-LC3 or anti-actin. Actin was used as a loading control. Densitometric LC3-II/actin ratios are shown underneath the blot (A). (B) Quantification of LC3-II/actin ratios. Ratios were normalized to ratio of infected cells with BafA1. Data, mean ± SEM (n = 6 independent experiments), *P < 0.05 (paired t-test, infected vs. non-infected cells). (C) LC3 immunoblotting of THP-1 macrophages infected with M. smegmatis (M. sm) for 24 h at different MOI (0–25). Differentiated THP-1 cells were incubated with M. smegmatis for 2 h, washed, and then incubated for 24 h. Cells were treated with 100 nM Bafilomycin A1 or DMSO control during the last 2 h. Cells were lysed and analyzed by immunoblotting with anti-LC3 or anti-actin. Densitometric LC3-II/actin ratios are shown underneath the blot. (D) Quantitative real-time PCR analysis of autophagy-related gene transcripts upon M. smegmatis infection. Differentiated THP-1 cells were incubated (Msm) or not (NI) with M. smegmatis at MOI 10 for 2 h, washed and incubated for 6 h. After lysis, RNA was extracted and RT-PCR analysis was performed for selected autophagy pathway genes. Data are expressed as relative copy numbers (RCN) to GADPH (house-keeping gene). Data, mean ± SEM (n = 3 independent experiments), *P < 0.05, **P < 0.01 (unpaired t-test).