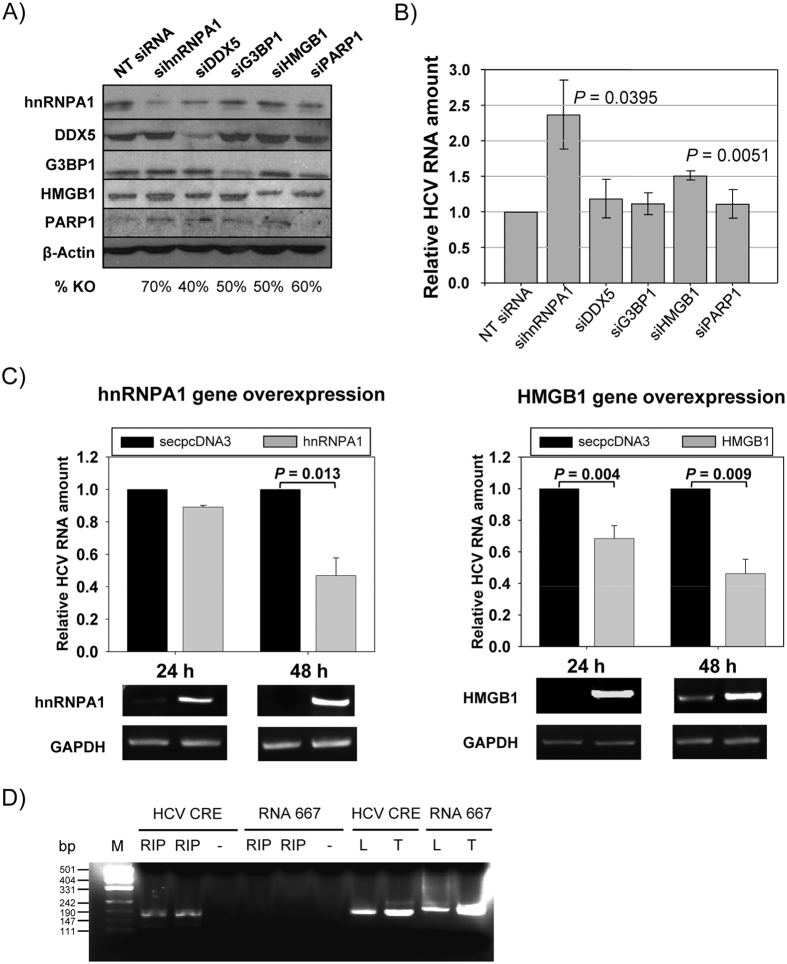
Figure 6. Effect of mRNA knockdown in HCV replication.
Replicon-containing cells (Huh-7-NS3-3′) were transfected with siRNAs directed against either hnRNPA1, DDX5, G3BP1, HMGB1 or PARP1, or with a non-targeting siRNA. After 48 h the cells were collected and RNA or protein extracted as described in Methods. (A) Protein lysates from Huh-7-NS3-3′cells transfected with siRNA were resolved by SDS-PAGE and immunoblotted with specific antibodies for the assayed proteins or for a β-actin control. (B) RNA lysates were retrotranscribed and cDNA corresponding to HCV replicon amplified by qPCR. Changes in replicon levels were represented as fold changes normalized to GAPDH. Standard deviation is displayed for each siRNA. Significant differences between siRNAs and non-targeting siRNA are indicated above the corresponding bar (p < 0.05 in a two-tailed Student’s t-test). (C) Overexpression of hnRNPA1 and HMGB1 coding genes. Replicon levels, normalized to GAPDH, are represented after 24 h and 48 h after transfection. P values indicating the statistical significance are included. Values are the mean of three independent experiments. A picture of a representative agarose gel showing the relative hnRNPA1 and HMGB1 mRNA levels to the GAPDH mRNA ones analyzed by RT-PCR is shown under each bar graph. (D) Immunoprecipitation of CRE and RNA 667 with anti-hnRNPA1 antibody. After RIP, samples were subjected to RT-PCR and resolved in a 2% agarose gel. Immunoprecipitated (RIP) samples from cells transfected with CRE and RNA 667 were run in duplicates with a negative sample without template (−). As positive controls, not immunoprecipitated lysates (L) and direct RNA template (T) were retrotranscribed and amplified. pUC19 plasmid digested with MspI was used as DNA size marker (M). Fragment sizes, in bp, are indicated on the left.