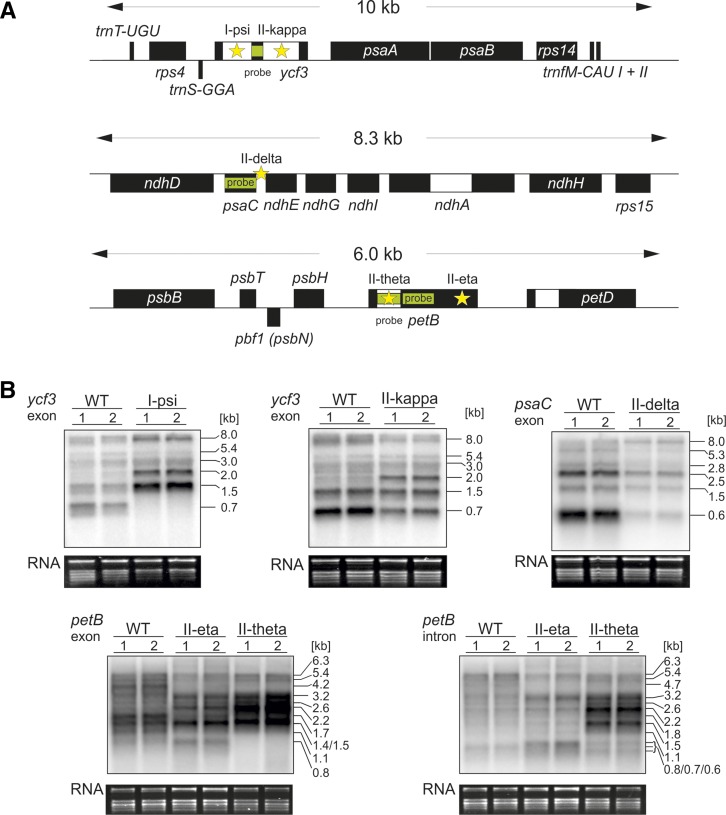
Figure 4.
Defects in mRNA Maturation Due to Mutations in Introns, a Coding Region, or an Intergenic Region in the Chloroplast Mutants I-psi, II-kappa, II-eta, II-theta, and II-delta.
(A) Schematic overview of the ycf3, psaC, and petB regions of the plastome. Positions of the mutations are marked with stars, and hybridization probes are indicated as green bars.
(B) RNA gel blot analyses of mRNA accumulation and RNA processing patterns. Upper row, left and middle panel: Splicing defects in ycf3 as found in the mutants I-psi and II-kappa, respectively. In I-psi, a 5-bp duplication in intron 1 leads to loss of splicing of this intron, correlating with a severe bleaching phenotype (Table 1). In II-kappa, a transversion in intron 2 results in a reduced splicing activity. Note that some mature transcript (0.7 kb) is still produced, consistent with the homoplasmic mutant being viable upon cultivation in soil (Table 1). Upper row, right panel: Processing defect in the psaC operon of the mutant II-delta. The mutant harbors a 24-bp deletion in the psaC-ndhE intergenic spacer. Whereas a precursor transcript of ∼8.0 kb overaccumulates in the mutant, accumulation of processed mRNA species is strongly reduced. Some mature monocistronic mRNA (0.6 kb) is produced in the mutant, consistent with its mild phenotype (Table 1). Lower row: Splicing and processing defects in transcripts from the psbB operon (containing the petB gene) in the mutants II-eta and II-theta. A 2-bp deletion is present in the petB intron in II-theta. In II-eta, a 5-bp duplication is present in the second exon of petB. Left panel: Hybridization to a petB exon probe; right panel: intron probe. In II-theta, a petB splicing defect is clearly visible. Transcripts overaccumulating in the mutant (i.e., the 2.2- and 1.5-kb RNA species) likely represent the unspliced petB/petD dicistron and the unspliced petB, respectively. The putative mature transcript (0.8 kb) does not accumulate in the mutant, in agreement with its severe phenotype (Table 1). Interestingly, in II-eta, the mutation in exon 2 leads to loss of the spliced psbH/petB/petD tricistron (1.7 kb in the wild type; Westhoff and Herrmann, 1988) and instead to accumulation of a 1.4-kb transcript likely containing the spliced petB/petD dicistron. The mature transcript (0.8 kb) seems to accumulate, which, however, carries a frameshift. This leads to a largely identical phenotype as II-theta (Table 1). For details on the mutations, see Table 1. 1 and 2 represent biological replicates of all plant lines analyzed here. To confirm equal loading, the ethidium bromide-stained agarose gel prior to blotting is also shown.