Figure 4C.
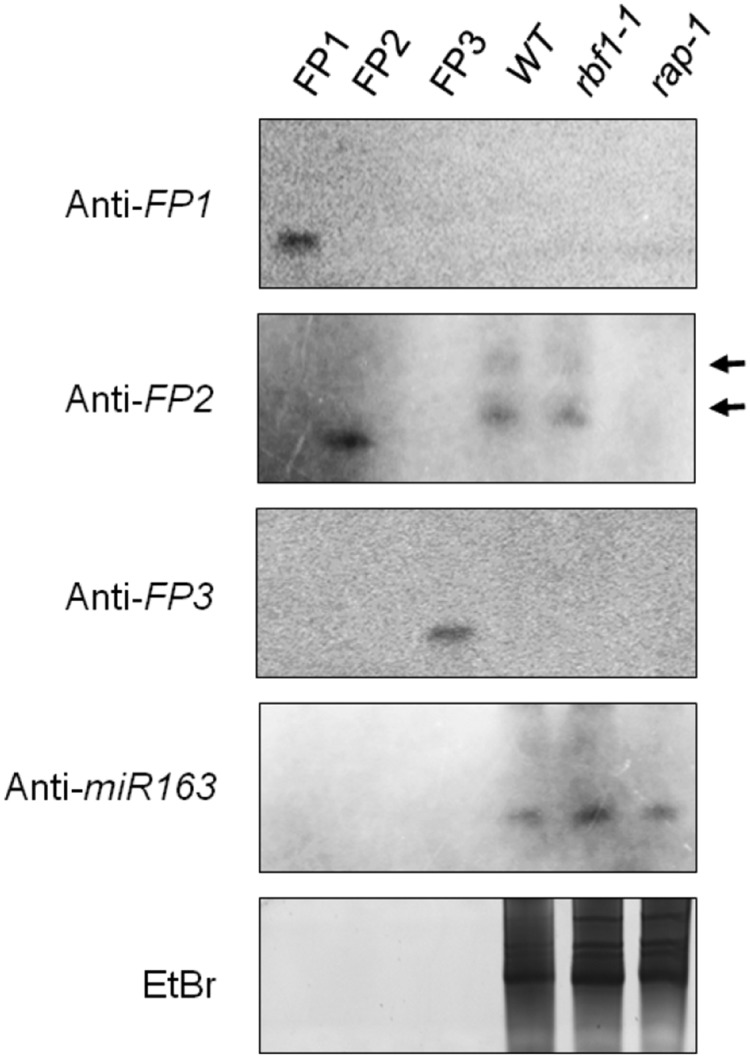
Corrected: Analysis of Small RNAs in rap-1.
RNA gel blot analyses of small RNAs from the wild type, rap-1, and an additional control RNA from the Arabidopsis rbf1-1 mutant, described to also reveal a defect in 16S rRNA processing (Fristedt et al., 2014). Thirty micrograms of total leaf RNA was fractionated in denaturing polyacrylamide gels and transferred to a charged nylon membrane. DNA oligonucleotides that mimic each sRNA (FP1-FP3) were run in adjacent lanes. DNA probes used, which are complementary to the respective small RNA, are indicated on the left. Note that single-stranded DNA migrates slightly faster than single-stranded RNA. As a positive control, we used microRNA miR163 (Ha et al., 2009). Two small RNAs specific to FP2 that were only detected in the wild type and rbf-1, but not in the rap-1 mutant, are indicated by black arrows. A representative ethidium bromide-stained gel is shown to demonstrate equal loading.
