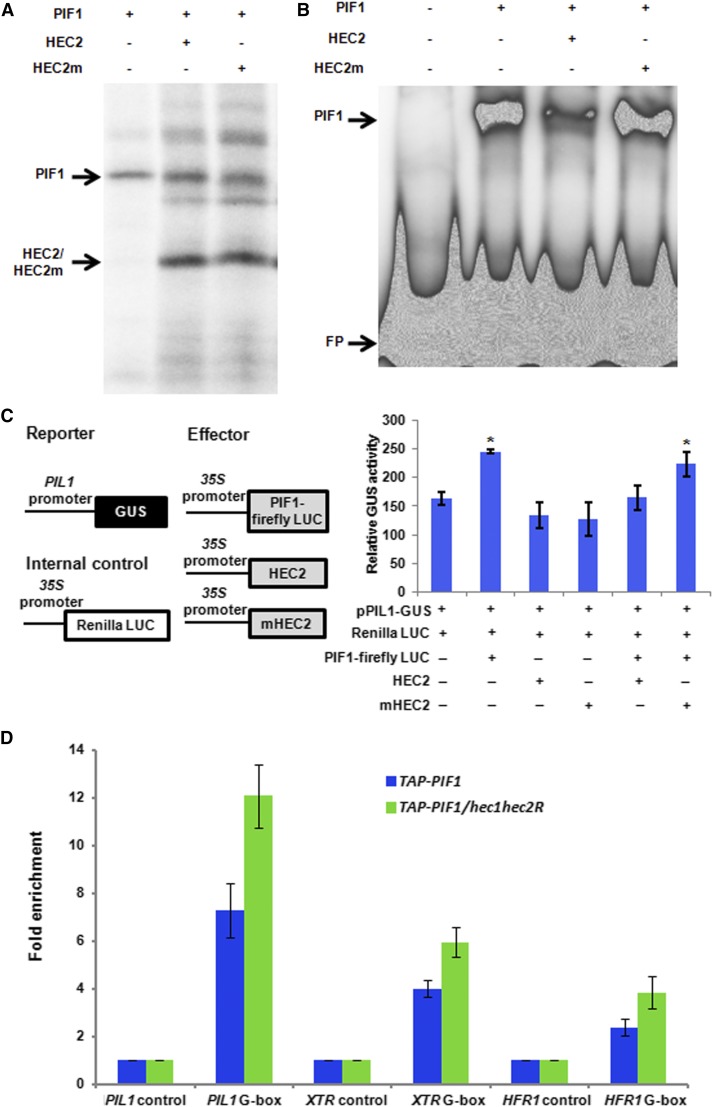
Figure 6.
HEC2 Inhibits the DNA Binding and Transcriptional Activation Activity of PIF1.
(A) A gel photograph shows the amount of protein used for the EMSAs. PIF1 and the wild type or mutant form of HEC2 clones were coexpressed in TnT system with 35S-Met labeling.
(B) EMSA showing PIF1 binding to PORC G-box is inhibited by HEC2, but not by the mutant version of HEC2. The same amount of TnT mix shown in (A) without the 35S-Met labeling was used in EMSA. A total of 30,000 cpm of labeled probe was used in each lane. EMSA conditions are as described (Moon et al., 2008).
(C) HEC2 inhibits the transcriptional activation activity of PIF1. Left: Schematic illustration of reporter, effectors, and internal control constructs used in transient promoter activation assay. Right: 2.5-d-old pif1-2 seedlings were transiently transformed with the different combinations of constructs indicated below. Relative expression of GUS activity was measured. The data were normalized by protein concentration and Renilla luciferase activity. The GUS activities of the control group, which was transiently expressed pPIL1:GUS and 35S:Renilla Luciferase, are set to 1 (n = 3 trials, each with four technical replicates, ±se, P < 0.01).
(D) HEC2 inhibits the promoter occupancy of PIF1 in vivo. ChIP assays show TAP-PIF1 binding to the G-box motif of PIF1 target promoters. The ChIP assay was performed on 3-d-old dark-grown seedlings expressing the TAP-PIF1 fusion protein either in pif1-2 or pif1-2 hec1 hec2RNAi line. Anti-MYC antibodies were used to immunoprecipitate TAP-PIF1 and associated DNA fragment. DNA was amplified using primers specific to the G-box fragments or control regions in PIL1, XTR, and HFR1 promoters.