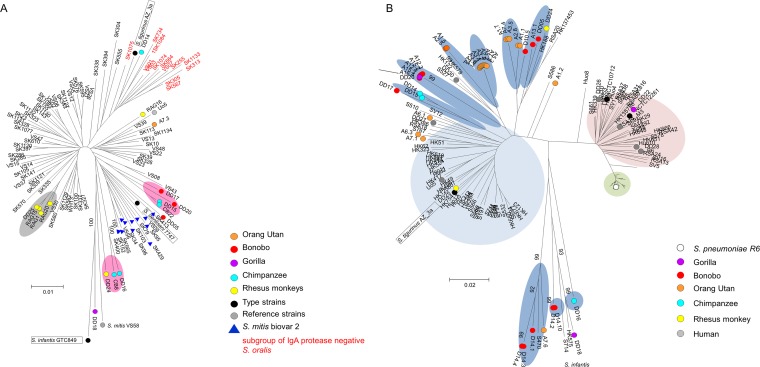
FIG 2 .
Phylogenetic trees of S. oralis and S. mitis strains. (A) Tree generated by MLSA loci of S. oralis from this study and sequences of S. oralis strains, S. tigurinus AZ_3a, and S. dentisani 7747 from the MLSA tree shown in Fig. 1. Gray shading, cluster of isolates from rhesus monkeys; pink shading, lineages consisting of primate isolates only. One S. mitis strain from the study of Bishop et al. (8), VS58, was included for comparison. Red letters indicate the subgroup of IgA protease-negative S. oralis isolates specified in reference 8. Bootstrap values (percentages) are based on 1,000 replications. (B) Phylogenetic relationship of primate isolates, including 38 isolates from zoo animals determined by MLST (7) but excluding ddl, combined with sequences of 119 human isolates (S. oralis, S. mitis, and S. pneumoniae) from different geographic locations (10) (isolate numbers are preceded by Hu for Hungary, RSA for South Africa, S for Spain, HK for Hong Kong, and B for Germany). Lineages within the S. oralis cluster containing only isolates from primates are shaded in dark blue. Bootstrap values (percentages) are based on 500 replications. The bar refers to genetic divergence as calculated by the MEGA software.