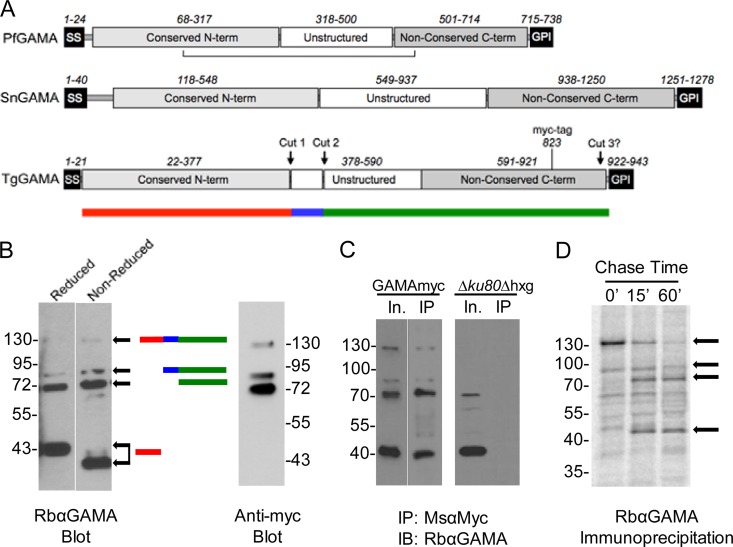
FIG 2 .
Domain organization of GAMA proteins and proteolytic processing of TgGAMA. (A) Schematic representations of PfGAMA, SnGAMA, and TgGAMA. Numbers depict approximate amino acid positions of major domains or elements. The horizontal line below PfGAMA indicates an intradomain disulfide bond. Approximate proteolytic processing sites are shown for TgGAMA. The position of a myc tag upstream of the GPI anchor at amino acid 823 in TgGAMA is shown. The colored bar below TgGAMA symbolizes junctions of the processing sites for panel B. term, terminus. (B) Western blot assay (left) of reduced and nonreduced parasite lysates with color bars on the right indicating the predicted fragments. Western blot assay (right) probed with anti-Myc antibody recognizes only the full-length protein and C-terminal fragments. (C) Parasite lysate input (In.) immunoprecipitated (IP) with mouse anti-Myc antibody and probed with rabbit anti-GAMA antibody (GAMAmyc lysate, left blot; Δku80 lysate, right blot). IB, immunoblot. (D) 35S metabolic labeling, followed by immunoprecipitation with anti-GAMA. Arrows indicate GAMA-specific bands based on Western blotting patterns. The values beside the blots in panels B to D are molecular sizes in kilodaltons.