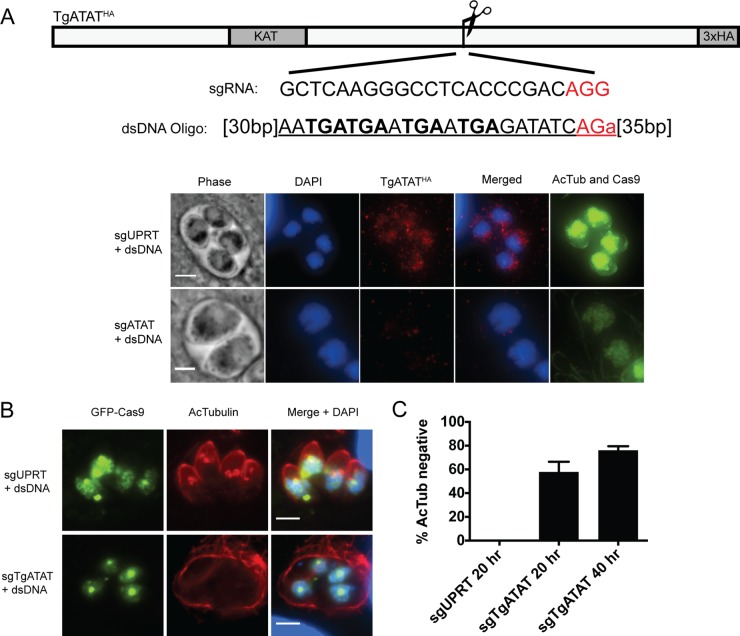
FIG 5 .
Selective targeting of GFP-Cas9 to the TgATAT locus eliminates K40 acetylation. (A) Diagram showing the site on TgATAT targeted by GFP-Cas9. The 20-bp TgATAT sgRNA sequence is shown; it is immediately upstream of the protospacer adjacent motif sequence (PAM, red). The dsDNA oligomer used for recombination is shown, and in brackets are the numbers of bases of homology flanking the PAM site. Underlined is the exogenous sequence introduced, including the four stop codons in bold. IFAs of dividing parasites expressing GFP-Cas9 confirm that disruption of TgATAT and loss of K40 acetylation occur only when Cas9 is targeted to the TgATAT locus (sgTgATAT). (B) IFA of TgATATHA parasites 40 h posttransfection with dsDNA oligomers and GFP-Cas9 targeted to either the UPRT (sgUPRT) or the TgATAT (sgTgATAT) locus. Scale bar, 3 μm. (C) Bar graph showing the percentage of GFP-Cas9-positive vacuoles that are acetyl-K40 negative in parasites in which GFP-Cas9 was targeted to UPRT (sgUPRT) versus TgATAT (sgTgATAT) 20 or 40 h after transfection. Error bars show the standard errors of the means (n = 3).