Fig. 3. A mutant screen to identify genes involved in chloroplast stress and degradation.
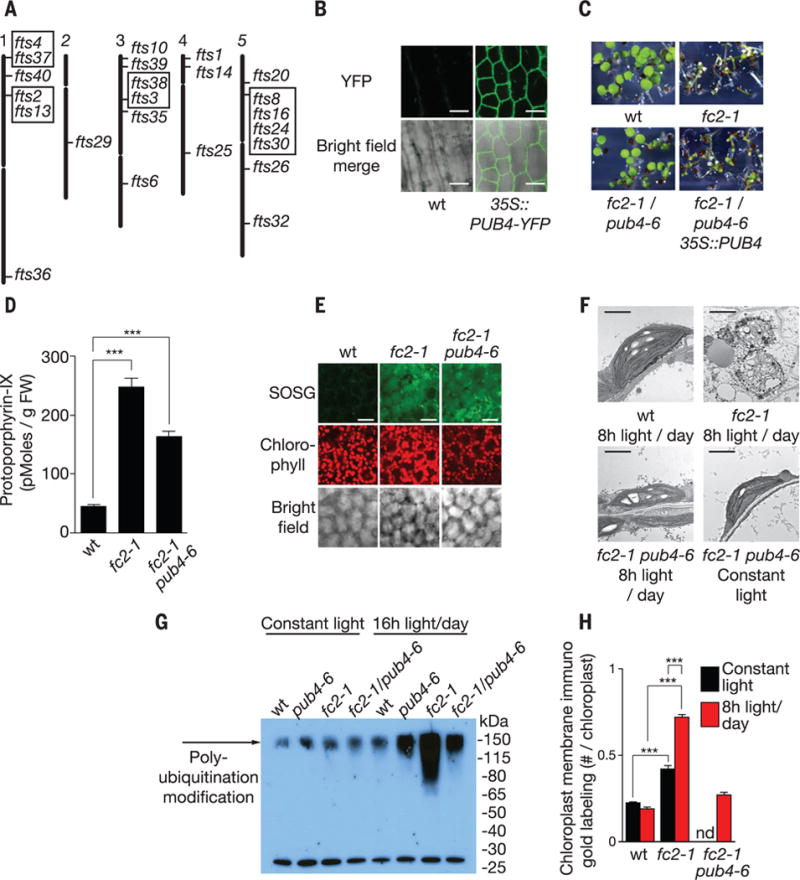
(A) Locations of 24 independent fts (ferrochelatase two suppressors) mutations representing 17 loci. Boxed mutants are allelic. (B) Subcellular localization of stably expressed PUB4-YFP in hypocotyl cells. (C) Complementation of the fts29 (pub4-6) phenotype with a wt copy of PUB4 grown for 5 days in 8 hours of light/day. (D) Proto levels in 3-day-old seedlings provided 8 hours of light/day 10 min after dawn (n = 3 replicates). (E) Representative confocal images of 1O2 accumulation in cotyledon mesophyll cells (2 hours after dawn) of 3-day-old seedlings provided with 8 hours of light/day. Scale bars, 20 μm. (F) Representative TEM micrographs of chloroplasts in 3-day-old cotyledon mesophyll cells 6 hours after dawn. Scale bars, 2 μm. (G) Antiubiquitin immunoblot of whole chloroplast fractions isolated from leaves 3 hours after dawn. Plants were grown for 2 weeks in constant light and then transferred to 16 hours of light/day or kept in constant light for 2 days. (H) Quantification of chloroplast envelope-associated ubiquitin by immunoelectron microscopy (gold labeling) (19) in 3-day-old seedlings (n = 3 seedlings, 10 cells each) 1 hour after dawn. All values are means T SEM. ***P ≤ 0.001, two-tailed Student’s t test.
