Figure 2.
Somatic NEK9 Mutations Cause Nevus Comedonicus
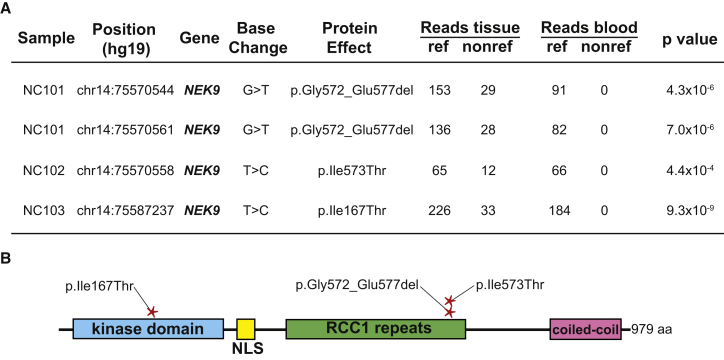
Whole-exome sequencing of DNA isolated from tissue and blood of individuals NC101, NC102, and NC103 was performed (A). Using MuTect, Perl and Python scripts, we identified tissue-specific SSNVs and indels and ranked them by Fisher’s exact test score. NC101 had two mutations in NEK9 that were in cis on spanning short reads. These mutations led to a loss of six amino acids in the encoded protein, p.Gly572_Glu577del, via abolition of the native exon 14 splice donor and utilization of a cryptic splice donor within the coding region of exon 14 (Figure S3). NC102 had a somatic NEK9 p.Ile573Thr missense variant. NC103 showed a somatic NEK9 p.Ile167Thr missense variant. No other somatic mutations were found in any of the three individuals with a Fisher’s exact test p value < 1 × 10−4, and no non-reference reads were identified in blood DNA in any individual. Mutations were confirmed with Sanger sequencing (Figure S2). NC101’s and NC102’s mutations were localized in the RCC1 domain of NEK9, whereas NC103’s mutation was localized in the kinase domain (B).