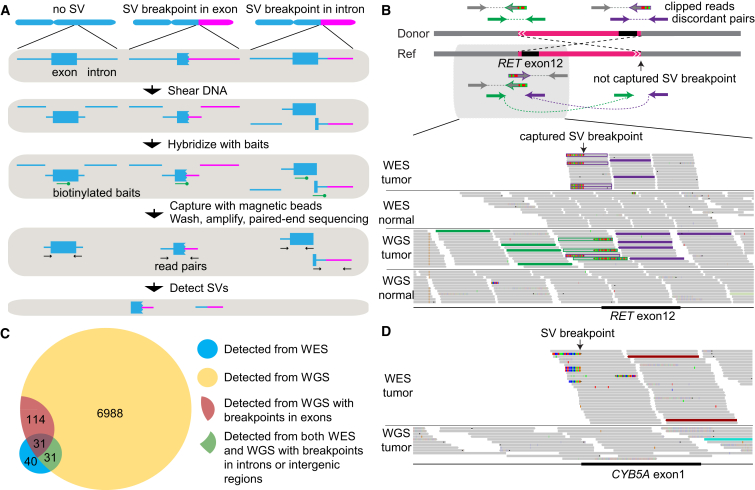
Figure 1.
Detecting Somatic SVs from WES Data
(A) Workflow showing how DNA fragments are captured and sequenced when SV breakpoints occur in exons and near exon-intron boundaries.
(B) A true somatic CCDC6-RET fusion resulting from a balanced inversion (chr10:61,655,977–43,611,997) in a thyroid cancer (TCGA-FK-A3SE) is detected by both WES and WGS. The scheme of the inversion is shown on the top (not to scale). The Integrated Genome Viewer screen shot for the captured breakpoint is shown on the bottom. Green and purple read pairs represent discordant pairs from two different breakpoints; one breakpoint is captured by WES and the other is not. The gray reads are concordant read pairs. The half-gray and half-striped reads with green or purple outlines are partially aligned (clipped) reads spanning the breakpoints.
(C) A Venn diagram showing the overlap between somatic SVs called from WES and WGS data.
(D) A true somatic deletion (chr18:71,930,713–71,958,983) in a lung adenocarcinoma (TCGA-91-6840) is detected by WES but not by WGS and is validated by PCR. The coverage in WES is >100× and there are six discordant read pairs (two displayed), whereas the coverage of WGS in the same region is 30× and no discordant read pair is present. The red reads are discordant read pairs supporting the somatic deletion.