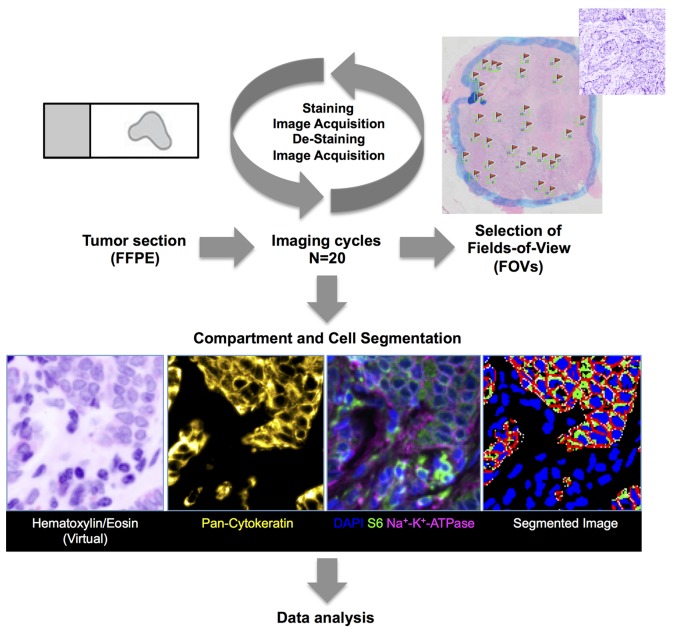
Figure 1. Experimental design.
Immunofluorescence approach. A single 3- to 5-μm unstained section from a routinely collected formalin-fixed and paraffin-embedded (FFPE) tumor tissue block was used from each tumor for the multiplex iterative imaging cycles (n = 20). Background autofluorescence (AF) tissue images were acquired before subsequent application of fluorescent dye-conjugated primary antibodies. Stained images were then acquired, followed by dye inactivation and restaining with new directly conjugated antibodies. New images were acquired, and the cycle was repeated until all target antigens were exhausted. Stained images were registered. Background AF was removed from each stained image. Images were segmented into epithelial and stromal regions using boundaries of cytokeratin staining, followed by identification of individual cells and corresponding plasma membrane (as determined by Na+K+ATPase staining), cytoplasm (S6 staining), and nuclear regions (DAPI). Biomarker pixel-level intensity data, which were subsequently queried in data analysis, were quantified at cell level. Three different metrics per marker (mean, standard deviation, and 90% hot spot) were used, amounting to about 155 million measurements. Data analysis included K-median clustering to groups of cells based on similar biomarker intensity levels. Each field of view (FOV) was manually reviewed. Only FOVs with >90% IDC cells on histopathological review were included in the analysis.