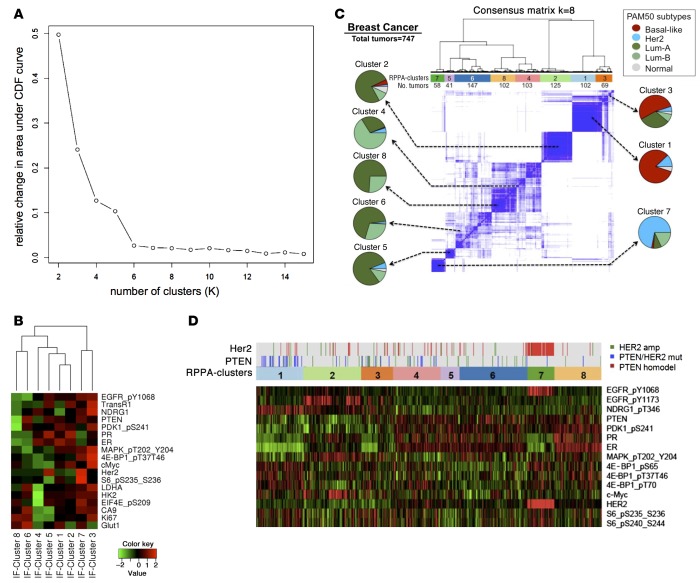
Figure 3. Protein coexpression clusters in human breast cancer.
(A) Consensus clustering was used to determine the optimum cluster set. The graph shows the relative change in area under cumulative distribution function for K = 2–15: when K increases, a positive change in the area under the curve decreases. After K = 8, the relative change is minimal and the curve flattens, suggesting that K = 8 is the best close-to-true partition. (B) Heatmap showing expression levels of each protein (log2) within the 8 clusters. IF, immunofluorescence. Rows were arranged following the lateral dendrogram. (C) Consensus hierarchical clustering of protein coexpression patterns in the TCGA breast cancer data set (747 human breast cancers). Protein expression was measured by reverse phase protein array (RPPA) using 187 antibodies. Pie charts indicate the association of each RPPA cluster with intrinsic breast cancer subtypes. Subtype annotation (PAM50) was available for 633 of 747 tumors. Numbers below each cluster indicate the number of human breast cancer samples belonging to each cluster. (D) Heatmap of the expression levels (RPPA) of the 10 proteins that were also represented in the multiplexed IF assay (see B). Genomic alterations in PTEN and HER2 are indicated above the RPPA cluster assignment.