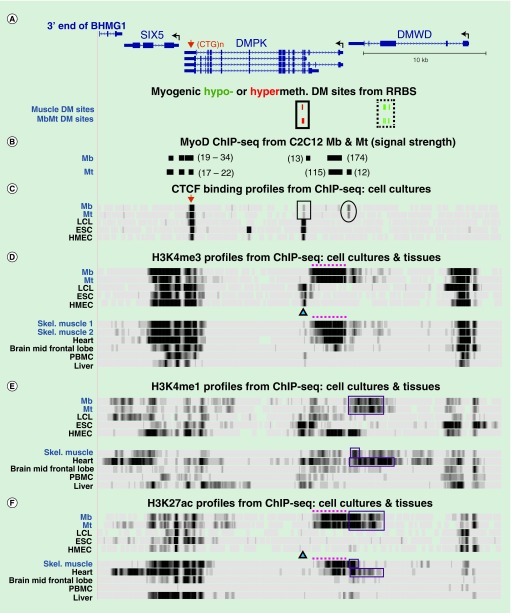
Figure 4. . Tissue-specific histone methylation and acetylation in the vicinity of DMPK.
(A) DMPK (four isoforms are shown), SIX5 and DMWD with myogenic DM sites indicated below (chr19:46,265,940–46,298,675). (B) Human DNA sequences orthologous to MyoD binding sites as deduced by MyoD ChIP-seq on murine C12C12 Mb and Mt [47]. Their relative signal strength in the C2C12 ChIP-seq is shown in parentheses. (C) CTCF ChIP-seq with a vertical viewing range of 0–50. Boxed region, tissue-specific CTCF site that displayed low signal in Mb and Mt and is adjacent to the myogenic hypermethylated DMR in DMPK; oval, CTCF site present preferentially in Mb and Mt. (C–F) ChIP-seq profiles for H3 methylation or acetylation as indicated. Triangles, position of the MbMt and skeletal muscle hypermethylated sites in DMPK; dotted lines and boxes, promoter-like and enhancer-like histone modifications, respectively, seen preferentially in Mb, Mt, skeletal muscle tissue and heart. CTCF ChIP-seq data for more samples are given in Supplementary Figure 1. DM: Differentially methylated; ESC: Embryonic stem cell; HMEC: Human mammary epithelial cell; LCL: Lymphoblastoid cell line; Mb: Myoblast; Mt: Myotube; NHLF: normal human lung fibroblasts; PBMC: Peripheral blood mononuclear cell.