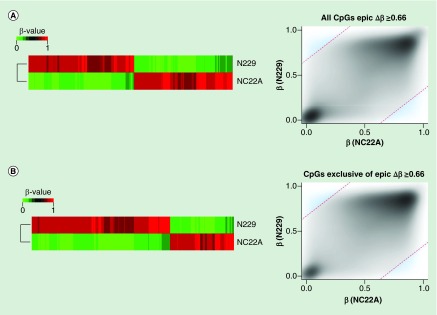
Figure 5. . Biological validation of the 850K DNA methylation microarray.
(A) Differentially methylated CpG sites (Δβ ≥0.66 ) on MethylationEPIC BeadChip microarray from a normal colon sample (NC22A), and normal sorted brain neurons (N229). Heatmap representation of differentially methylated CpG sites (left) and methylation values distribution (right) of samples where methylation differences threshold has been denoted as a discontinued red line (Δβ ≥0.66), and those values considered as differentially methylated have been highlighted in blue. (B) Differentially methylated CpG sites (Δβ ≥0.66), among the newly added 413,759 CpG sites of the MethylationEPIC BeadChip microarray, for a normal colon sample (NC22A), and normal sorted brain neurons (N229). Heatmap representation of differentially methylated CpG sites (left) and methylation values distribution (right) of samples where methylation differences threshold has been denoted as a discontinued red line (Δβ ≥0.66), and those values considered as differentially methylated have been highlighted in blue.