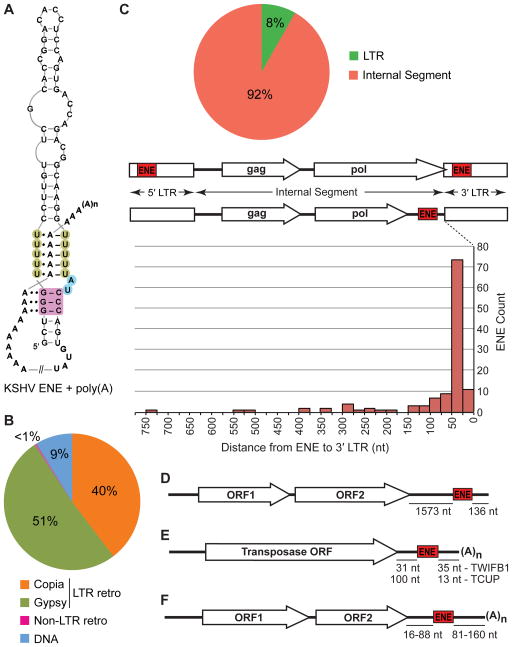
Figure 1. Distribution of the ENEs described in this paper between different TE classes and their positions within TEs and viral genomic RNAs.
(A) Interaction of the KSHV PAN RNA ENE with the poly(A) tail (Conrad et al., 2006; Mitton-Fry et al., 2010). The U residues involved in triple-helix formation are shaded green while the A-minor receptor pairs are colored magenta. Linker residues are shaded blue. Watson-Crick and Hoogsteen base pairings are denoted by dashes and single dots, respectively, while A-minor interactions are indicated by double dots. (B) Distribution of ENEs among different classes of TEs. (C) ENE positions within LTR retrotransposons. The pie chart depicts the partitioning of ENEs between LTRs and Internal Segments. The graph shows distances to the 3′ LTR for ENEs located in Internal Segments. The distances to the poly(A) tail of transcripts cannot be predicted because the sequences around the LTR polyadenylation sites lack conserved motifs. (D) The ENE of the non-LTR retrotransposon Tad1 from N. crassa is predicted to lie ~136 nt upstream of the poly(A) tail of the Tad1 RNA. (E) Positions of the TWIFB1_OSa and TCUP_ZMa ENEs in their respective hAT transposase mRNAs. (F) Location of ENEs in Dicistrovirus genomes. Distance ranges for MCDiV, TSV, MRTV and HiPV are shown (see Figure S3 for details). Not to scale.