SUMMARY
The ability of cancer cells to establish lethal metastatic lesions requires the survival and expansion of single cancer cells at distant sites. The factors controlling the clonal growth ability of individual cancer cells remain poorly understood. Here we show that high expression of the transcription factor ARNTL2 predicts poor lung adenocarcinoma patient outcome. Arntl2 is required for metastatic ability in vivo and clonal growth in cell culture. Arntl2 drives metastatic self-sufficiency by orchestrating the expression of a complex pro-metastatic secretome. We identify Clock as an Arntl2 partner and functionally validate the matricellular protein Smoc2 as a pro-metastatic secreted factor. These findings shed light on the molecular mechanisms that enable single cancer cells to form allochthonous tumors in foreign tissue environments.
Graphical abstract

INTRODUCTION
Numerous physical and biological constraints prevent normal epithelial cells and benign tumor cells from populating secondary sites, but metastatic cancer cells are able to overcome all of these hurdles to form secondary tumors in distant and diverse organ environments (Fidler, 2003; Nguyen et al., 2009a). The metastatic process is a highly inefficient process that depends on the ability of single cancer cells to survive transit through the circulatory system and enter and expand in a foreign site. The molecular basis of how individual cancer cells acquire the metastatic self-sufficiency is a fascinating yet poorly understood biological question of immense clinical importance.
Most cancer deaths are related to complications of metastatic growth and metastatic lung cancer is the leading cause of cancer deaths in both men and women in the United States, with over 155,000 patients dying each year in this country alone (Siegel et al., 2016). The mutations and DNA copy number alterations that drive lung cancer initiation and growth are now understood in increasing detail (The Cancer Genome Atlas Research Network, 2014). However, metastatic ability is unlikely to be fully explained by these genomic alterations, and there is a growing appreciation for the importance of transcriptional changes driven by lineage-specific and developmental transcription factors (Cheung and Nguyen, 2015; Ell and Kang, 2013). Lung adenocarcinoma is a major subtype of lung cancer, and although both pro- and anti-metastatic transcription factors that regulate distinct metastatic phenotypes have been recently identified, these few factors are unlikely to drive the entire compendium of malignant phenotypes required for optimal metastatic fitness (Cheung et al., 2013; Li et al., 2015, 2014; Nguyen et al., 2009b; Winslow et al., 2011).
Cancer cells within primary and metastatic tumors also integrate positive and negative cues from secreted factors and interact with a complex and dynamic microenvironment that includes the extracellular matrix (ECM) as well as diverse stromal cell populations. While secreted factors produced by cancer cells recruit stromal cells, dampen anti-tumor immunity, and promote ECM deposition and remodeling, they also function in an autocrine manner to drive resistance to targeted therapies, promote cell growth, and control cancer cell differentiation state (Bafico et al., 2004; Lu et al., 2003; Mo et al., 2013; Wilson et al., 2011; Zhu et al., 2014). Malignant progression and metastasis represent a multistep process, and each step is likely influenced by the ability of the cancer cells to create, modify, and interact with, their microenvironment.
The cancer cell secretome (including secreted proteins, lipids, exosomes, and other small molecular messengers) has been shown to have both short-range and systemic effects (Cox et al., 2015; McAllister and Weinberg, 2014). These secreted factors and their effects on cancer cells are likely cancer type-specific and dynamically remodeled during cancer progression, metastasis, and in response to therapy (Blanco et al., 2012; Obenauf et al., 2015). Secreted proteins are diverse and pleiotropic, including cytokines, growth factors, shed receptors, proteases, structural ECM components, and matricellular proteins. Matricellular proteins are a class of non-structural ECM proteins that modulate cell-matrix interactions and cellular functions such as adhesion or migration (Murphy-Ullrich and Sage, 2014). Most of the work demonstrating a role for secreted factors in metastatic ability has focused on breast cancer, where several factors have been shown to have a dramatic impact on different steps of cancer progression, metastasis, and therapeutic resistance (Cox et al., 2015; Korpal et al., 2011; McAllister and Weinberg, 2014; Obenauf et al., 2015; Oskarsson et al., 2011).
To form overt tissue-destructive metastases, disseminated cancer cells in secondary tissue environments must possess the ability to expand in that foreign site. The requirement for single cancer cells to survive and expand within a secondary site necessitates the acquisition of particular characteristics that neither normal cells nor benign cancer cells possess. These cell state alterations are likely driven by concerted changes in gene expression programs, but there is an incomplete understanding of the transcriptional networks that control lung cancer metastatic ability and almost no knowledge on whether remodeling of the cancer cell secretome promotes metastatic seeding and expansion. The transcription factor Arntl2 (Bmal2/Mop9/Clif) is a paralog of the circadian transcription factor Arntl (Bmal1/Mop3) (Hogenesch et al., 2000). While Arntl2 expression has been investigated in normal tissues and several tumor types, there have been no studies on its function in cancer or metastatic progression (Hogenesch et al., 2000; Mazzoccoli et al., 2012; Shi et al., 2010). Here we investigate a role for Arntl2 as a driver of lung adenocarcinoma metastatic ability.
RESULTS
Arntl2 is highly expressed in lung adenocarcinoma metastases
We previously performed global gene expression profiling on a panel of cell lines derived from primary tumors and metastases that developed after lentiviral-Cre infection of the lung epithelial cells of KrasLSL-G12D/+;Trp53flox/flox (KP) mice (Winslow et al., 2011). From this analysis, many genes of potential functional importance were identified, including the transcription factor Arntl2. Arntl2 is a basic helix-loop-helix (bHLH) factor that is more highly expressed in cell lines derived from metastatic primary lung tumors (T Met) and metastases (Met) than in cell lines derived from non-metastatic primary lung tumors (TnonMet; Figure 1A). We directly assessed the expression of Arntl2 in neoplastic cells with different genotypes and at different stages of malignant progression by isolating Tomatopos cells from tumors and metastases in KrasLSL-G12D/+;Trp53flox/flox;R26LSL-Tom (KPT) mice as well as from tumors in KrasLSL-G12D/+;R26LSL-Tom (KT) mice (Caswell et al., 2014). We confirmed that Arntl2 is highly expressed in metastatic primary tumors, disseminated tumor cells (DTCs) in the pleural cavity, and lymph node and distant metastases (Figure 1B).
Figure 1. Arntl2 is up-regulated in metastatic lung adenocarcinoma and high expression predicts poor lung adenocarcinoma patient outcome.
A. Gene expression of Arntl2 in cell lines derived from metastatic primary lung tumors (T Met) and metastases (Met) relative to cell lines derived from non-metastatic primary lung tumors (TnonMet) from the KrasLSL-G12D/+;Trp53flox/flox (KP) mouse model. Each dot represents a cell line and the bar is the mean. * p value <0.05.
B. qRT-PCR gene expression analysis for Arntl2 on purified neoplastic cells from defined genotypes and stages of cancer progression. KT tumors were taken from Trp53-sufficient KrasLSL-G12D/+;R26LSL-Tom mice. Expression is normalized to house keeping genes with the average of the TnonMet samples set to 1.
C. Analysis of TCGA lung adenocarcinoma data to uncover the ability of each individual gene to predict patient outcome. Each dot is a gene and ARNTL2 is indicated. For genes on the upper left high expression predicts shorter survival. For genes on the upper right high expression predicts longer survival.
D. ARNTL2 expression in human lung adenocarcinomas. Data from TCGA. The highest 10% (n=44) of patients are included. Hazard ratios (HR) and p values are shown. Median survival is indicated.
E. Survival of Stage I human lung adenocarcinoma patients with high versus low ARNTL2 expression. Meta-analysis data from Gyorffy et al.
F. Survival of human lung adenocarcinoma patients with high versus low ARNTL2 expressing tumors. Data from Shedden et al.
High ARNTL2 expression predicts poor lung adenocarcinoma patient survival
In addition to our global analysis of autochthonous mouse models of human lung adenocarcinoma, we also analyzed human lung adenocarcinoma gene expression datasets to determine whether ARNTL2 expression predicts patient outcome. We used TCGA gene expression and clinical data to determine how well the expression of each individual gene predicts lung adenocarcinoma patient outcome (Table S1). We determined the expression level of each gene that resulted in the most significant difference in survival and calculated the adjusted p value and hazard ratio for each gene. Given how little is known about ARNTL2 in cancer, we were surprised to find that high ARNTL2 expression is one of the strongest single gene predictors of lung adenocarcinoma patient survival (Figure 1C and Table S1). Across the >400 lung adenocarcinoma patients within TCGA, those with the highest ARNTL2 expression had significantly shorter survival related to cancer recurrence (Figure 1D–F). Patients whose tumors were within the highest decile of ARNTL2 expression had more than three times shorter median survival than all other patients combined (Figure 1D). When the analysis was limited to only patients that presented without lymph node metastasis (N0) and without residual tumor after resection (R0), high ARNTL2 expression was still a strong single gene predictor of poor patient outcome (Figure S1A and S1B). Finally, high ARNTL2 also predicted poor outcome for patients with oncogenic KRAS mutations (Figure S1C). In additional gene expression datasets, ARNTL2 expression consistently predicted poor lung adenocarcinoma patient outcome, including in Stage I patients within a large lung adenocarcinoma meta-analysis and in the Director’s Challenge microarray-based gene expression profiling dataset (Figure S1D–F) (Gy3rffy et al., 2013; Shedden et al., 2008).
ARNTL2 is expressed in poorly differentiated lung adenocarcinomas
Metastases that develop in the oncogenic Kras-driven, p53-deficient lung adenocarcinoma mouse model are poorly differentiated and lack the expression of lung differentiation markers, including the transcription factor Nkx2-1/Ttf-1 (Jackson et al., 2005; Winslow et al., 2011). Nkx2-1 and Arntl2 are expressed in a mutually exclusive manner in cancer cells isolated directly from tumors and metastases, as well as in cell lines derived from these distinct cancer cell states (Figure 2A, 2B and S2A).
Figure 2. ARNTL2 expression is associated with poorly differentiated lung adenocarcinomas.
A. qRT-PCR for Nkx2-1 and Arntl2 on ex vivo sorted cancer cells from KPT tumors and metastases. Note split scale for Arntl2.
B. Western blot for Arntl2 on ex vivo sorted cancer cells from KPT mice. Representative of five metastases (Met) from three KPT mice. Actin shows loading.
C. Expression of ARNTL2 and the lung differentiation marker NKX2-1/TTF1 in human lung adenocarcinoma. Each dot represents a lung adenocarcinoma from TCGA (n=230).
D. ARTNL2 expression in human lung adenocarcinomas. Data from Shedden et al. The highest 20% of all samples are defined as ARNTL2high. Number of ARNTL2high/total tumors is shown. Percent of ARNTL2high tumors is shown in brackets.
E. RNAscope-based ARNTL2 expression in human lung adenocarcinoma cells at low (top panel) and high (bottom panel) magnification. Hematoxylin counter-stained well-differentiated adenocarcinoma (left) and poorly differentiated adenocarcinoma (right) are shown. Top scale bars = 50 μm. Bottom scale bars = 25 μm.
F. Quantification of ARNTL2 expression relative to cytological differentiation state. The highest 20% of all samples are defined as ARNTL2high. Number of ARNTL2high/total tumors is shown. Percent of ARNTL2high tumors is shown in brackets.
See also Figure S2.
Within human lung adenocarcinoma gene expression profiling datasets, canonical genes associated with lung adenocarcinoma differentiation were consistently among the most negatively correlated with ARNTL2 (Figure 2C, S2B, and data not shown). ARNTL2 was also less likely to be highly expressed in tumors that were pathologically graded as well-differentiated (Figure 2D).
To confirm that ARNTL2 is expressed within the cancer cells themselves in human lung adenocarcinoma and to further investigate the association of ARNTL2 with a differentiated state, we assessed ARNTL2 expression in human lung adenocarcinoma sections. We were unable to identify an anti-ARNTL2 antibody that worked for immunohistochemistry, therefore we performed RNA in situ hybridization for ARNTL2 on more than 150 human lung adenocarcinoma samples. We detected ARNTL2 expression specifically in the cancer cells and only in a subset of human lung adenocarcinomas. Consistent with the analysis of gene expression dataset, these ARNTL2positive tumors were enriched for those with moderate to poorly differentiated histology (Figure 2E, 2F, and S2C).
Arntl2 is required for lung adenocarcinoma metastasis
Our correlative data from mouse and human lung adenocarcinomas suggested that Arntl2 could be a pro-metastatic transcription factor. To determine whether Arntl2 is a functional regulator of metastatic ability, we initially knocked-down Arntl2 in a lung adenocarcinoma cell line derived from a lymph node metastasis from the KrasLSL-G12D/+;Trp53flox/flox model (482N1)(Winslow et al., 2011). Stable knock-down of all isoforms of Arntl2 with two distinct shRNAs was confirmed at the RNA and protein level (Figure 3A, 3B, and S3A). While Arntl2 knock-down had no effect on proliferation or cell death under standard culture conditions, 482N1shArntl2 cells formed many fewer lung and liver metastases after intravenous transplantation into syngeneic immunocompetent recipient mice (Figure S3B, S3C, and Figure 3C–E). 482N1shArntl2 cells also formed fewer liver metastases after intrasplenic injection, consistent with Arntl2 being a driver of multi-organ metastatic ability (Figure S3D and S3E).
Figure 3. Arntl2 is required for lung adenocarcinoma metastatic ability.
A. qPCR for Arntl2 expression in a metastasis-derived cell line (482N1) with or without knock-down of Arntl2 by two independent shRNAs, normalized to shControl and Gapdh. **** p value <0.0001.
B. Western blot for Arntl2 protein levels in 482N1 cell lines, with or without Arntl2 knock-down. Actin shows loading.
C. Representative histology of metastatic lung lesions three weeks after intravenous transplantation of 482N1 cell lines. Scale bars = 3 mm.
D. Lung weight per mouse, measured three weeks after intravenous transplantation of 482N1 cell lines. Each dot represents a mouse and the bar is the mean. Normal lung weight is indicated. * p value <0.05; p value for shControl versus shArntl2#2 = 0.0606.
E. Quantification of tumor area relative to total lung area. n > 4 mice/group. ** p value <0.005 ***; p value <0.001.
F. 889DTC subcutaneous tumor growth in immune compromised NSG mice with or without Arntl2 knock-down. Subcutaneous tumor mass from three to five mice/group is shown. NS = not significant.
G. Fluorescent dissecting scope images of whole lungs from immune compromised NSG mice with 889DTC subcutaneous tumors with or without Arntl2 knock-down (top). Representative histology images of lung lobes from the same NSG mice (bottom). Upper scale bars = 5 mm. Lower scale bars = 1 mm.
H, I. Quantification of lung and liver metastasis number by counting surface tumors. ** p value <0.01; *** p value <0.001.
Panels with error bars show the mean +/− SD. See also Figure S3.
To confirm the functional importance of Arntl2 on lung cancer metastatic ability, we knocked-down Arntl2 in a second murine cell line, 889, which was generated from disseminated tumor cells (DTC) from the pleural cavity of a lung adenocarcinoma-bearing KPT mouse. Subcutaneous growth of 889 cells in immunocompromised NSG recipient mice leads to the formation of spontaneous metastases in the liver and lung. Arntl2 knock-down did not change proliferation or cell death under standard culture conditions or alter subcutaneous tumor growth (Figure S3F–H). The subcutaneous tumors that formed from shArntl2 cells did not show any signs of acquiring epithelial differentiation, but Arntl2 knock-down greatly reduced both lung and liver metastases (Figure 3H and 3I). 889shArntl2 cells also seeded fewer metastases after intravenous transplantation, and the metastases that formed lacked Arntl2 knock-down (Figure S3I and S3J). These results further suggest that Arntl2 promotes lung adenocarcinoma metastatic seeding or outgrowth.
In human lung adenocarcinoma, NKX2-1 is one of the most anti-correlated genes with ARNTL2. NKX2-1 is a regulator of lung differentiation and a suppressor of metastatic ability (Kimura et al., 1996; Mu, 2013; Winslow et al., 2011), therefore we determined whether Nkx2-1 and Arntl2 regulate the expression of each other. Neither Nkx2-1 knock-down in a non-metastatic tumor-derived cell line nor Nkx2-1 re-expression in a metastatic tumor-derived cell line altered Arntl2 expression (Figure S2D–G). Arntl2 cDNA expression in a non-metastatic murine lung adenocarcinoma cell line did not reduce Nkx2-1 expression, and Arntl2 knock-down in metastasis-derived mouse cell lines and human lung adenocarcinoma cell lines did not de-repress Nkx2-1 (Figure S2H, S2I, and data not shown). Thus, while expression of these factors represent distinct cellular lung adenocarcinoma states, they do not regulate the expression of one another. High expression of the chromatin-associated protein Hmga2 also marks the poorly differentiated metastatic lung adenocarcinoma state, but Arntl2 and Hmga2 do not appear to regulate the expression of each other (Winslow et al, 2011 and Figure S2H, S2J–S2L).
Arntl2 specifically regulates the clonogenic ability of mouse lung adenocarcinoma cells
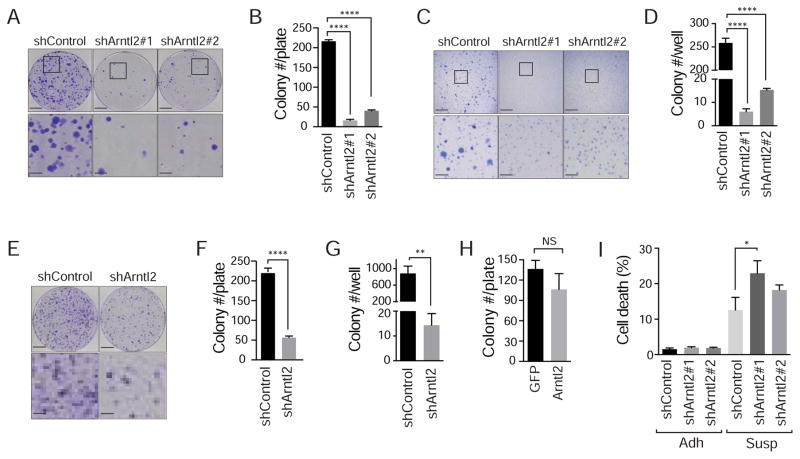
To better define the cellular phenotypes driven by Arntl2, we assessed the impact of Arntl2 knock-down on cell culture assays that test motility and invasion, anoikis resistance, clonal growth potential, and growth under anchorage-independent conditions. In addition to having no effect on proliferation and cell death under normal culture conditions (Figure S3A and S3B, and S3E–G), knock-down of Arntl2 in 482N1 and 889 cells had a minimal effect on migration and invasion (Figure S4A–B and data not shown). However, Arntl2 was critical for clonal growth ability. When 482N1 cells were plated in conditions where single cells had to expand into colonies (both in conditions of low-density plating and in anchorage-independent conditions in soft agar), Arntl2 knockdown reduced the number of colonies by more than five-fold (Figure 4A–D). These in vitro phenotypes were recapitulated in 889 cells with Arntl2 knock-down (Figure 4E–G and S4C). To determine whether Arntl2 is sufficient to increase clonal growth ability, we expressed Arntl2 in a cell line derived from a non-metastatic primary tumor (394T4; Figure S4D). Expression of exogenous Arntl2 did not increase clonal growth ability, suggesting that Arntl2 requires additional factors in the metastatic state to promote seeding efficiency and outgrowth (Figure 4H).
Figure 4. Arntl2 is required, but not sufficient for clonal expansion of lung adenocarcinoma cells.
A, B. 482N1 lung adenocarcinoma colony formation under low-density plating conditions (3x103 cells/10 cm plate) with or without Arntl2 knock-down. Upper scale bars = 2 cm, lower scale bars 5 mm. **** p value <0.0001.
C, D. 482N1 lung adenocarcinoma cell growth in anchorage-independent conditions (soft agar) with or without Arntl2 knock-down. Upper scale bars = 5 mm. Lower scale bars = 1.5 mm. **** p value <0.0001.
E, F. 889DTC lung adenocarcinoma colony formation under low-density plating conditions (3x103 cells/10 cm plate) with or without Arntl2 knock-down. Upper scale bars = 2 cm, lower scale bars 5 mm. **** p value <0.0001.
G. 889DTC lung adenocarcinoma cell growth in anchorage-independent conditions (soft agar) with or without Arntl2 knock-down. ** p value <0.01.
H. Quantification of colony formation in low-density plating conditions of 394T4 (TnonMet) cells with or without over-expression of Arntl2. NS = not significant.
I. Quantification of percent DAPIpositive dead cells after plating in adherent (Adh) and suspension (Susp) conditions of 482N1 cells with or without Arntl2 knock-down. * p value <0.05.
Panels with error bars show the mean +/− SD. See also Figure S4.
Anoikis, a form of cell death induced by lack of attachment, is a potential anti-metastatic mechanism that limits the ability of a cancer cell to survive in the circulation and initially within the secondary site. 482N1shArntl2 cells cultured in suspension conditions exhibited more cell death, suggesting that the reduced clonal growth ability could be partially driven by reduced resistance to anoikis (Figure 4I).
ARNTL2 promotes the clonogenic ability of human lung adenocarcinoma cells
To investigate the function of ARNTL2 in human lung cancer, we assessed the effect of ARNTL2 knock-down in two human lung adenocarcinoma cell lines: H1792 (Figure 5A–E) and H2009 (Figure 5F–I). Both H1792 and H2009 have oncogenic KRAS mutations (G12C and G12A, respectively) and express high levels of ARNTL2 (data not shown). H1792 was derived from a pleural effusion, while H2009 was derived from a lymph node metastasis. Knock-down of all ARNTL2 protein-coding isoforms in each cell line with two shRNAs was confirmed at the RNA and protein level (Figure 5A and 5F, S5A and S5I and S5J). Consistent with our results from the murine lung cancer cell lines, ARNTL2 knock-down in H1792 and H2009 cells did not significantly change proliferation or cell migration under standard culture conditions and did not induce any obvious changes in cell morphology (Figure 5B, 5G, S5B–S5D, S5F–S5H, and data not shown). ARNTL2 knock-down reduced the ability of H1792 cells to form colonies under low-density plating and anchorage-independent conditions, and H2009shARNTL2 cells also formed fewer colonies under low-density conditions (Figure 5C, 5D and 5H). In each human cell line, ARNTL2 knock-down also increased anoikis (Figure 5E and 5I).
Figure 5. ARNTL2 is required for clonogenic expansion of human lung adenocarcinoma cells.
A. qPCR for ARNTL2 expression in control and knock-down H1792 cells, normalized to ACTIN. **** p value <0.0001.
B. PrestoBlue assay for H1792 cell growth in standard culture conditions, with or without ARNTL2 knock-down.
C, D. H1792 colony formation in anchorage-independent conditions (C) and low-density plating conditions (D) with or without ARNTL2 knock-down. * p value <0.05; ** p value <0.01.
E. Quantification of percent DAPIpositive dead cells after plating in adherent (Adh) and suspension (Susp) conditions of H1792 cells with or without Arntl2 knock-down. *** p value <0.001.
F. qPCR for ARNTL2 expression in control and knock-down H2009 cells, normalized to ACTIN. *** p value <0.001; **** p value <0.0001.
G. PrestoBlue assay for H2009 cell growth in standard culture conditions with or without ARNTL2 knock-down.
H. H2009 colony formation under low-density plating conditions, with or without ARNTL2 knock-down. *** p value <0.001.
I. Quantification of percent DAPIpositive dead cells after plating in adherent (Adh) and suspension (Susp) conditions of H2009 cells with or without ARNTL2 knock-down. *** p value <0.001; * p value <0.05.
Panels with error bars show the mean +/− SD. See also Figure S5.
Collectively, our in vitro and in vivo data suggest that Arntl2 is particularly important for the ability of cancer cells to survive as single cells and to seed colonies that ultimately have the potential to form tissue-destructive metastases.
Arntl2 regulated genes are enriched for secreted factors
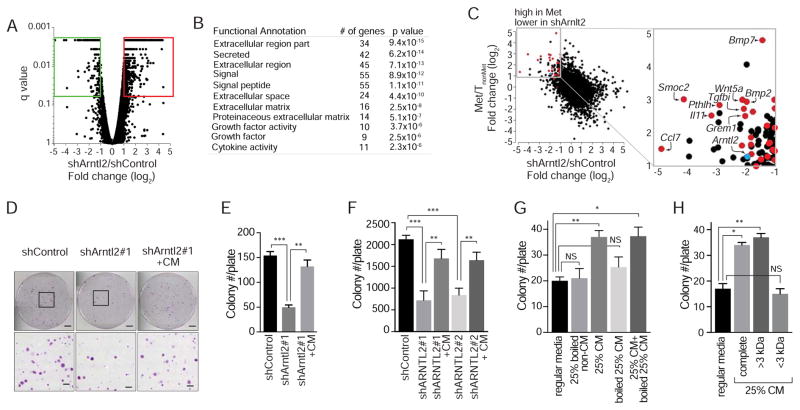
As an initial step toward understanding how Arntl2 drives metastatic ability, we performed RNA-seq-based gene expression profiling on metastatic lung cancer cells with and without Arntl2 knock-down. Surprisingly, the genes down-regulated in 482N1shArntl2 cells were highly enriched for secreted proteins, with some factors >10-fold reduced in shArntl2 cells (Figure 6A, 6B, and Figure S6). Many of these Arntl2-regulated secreted factors were also more highly expressed in metastasis-derived cells lines than in non-metastatic primary tumor-derived cell lines (Figure 6C). Several of these top secreted factors, including Wnt5a, Il-11, and Cxcl5 have been previously implicated in regulating metastatic ability in lung and other cancer types (Bo et al., 2013; Hanaki et al., 2012; Kang et al., 2013; Weeraratna et al., 2002; Zhou et al., 2014), while others, such as Smoc2, Grem1, and Ltbp2, are relatively poorly characterized in metastasis (Table S2 and Shvab et al., 2015). Arntl2 knock-down did not lead to changes in canonical epithelial genes Cdh1 and Epcam or lung differentiation genes Muc1 and Abca3 consistent with Arntl2 not directly affecting cancer cell differentiation (Table S2).
Figure 6. Arntl2-regulated secreted factors drive clonal growth ability.
A. RNA-seq on 482N1shControl and shArntl2 cells to identify differentially expressed genes. shArntl2 is an average of 482N1shArntl2#1 and 482N1shArntl2#2.
B. GO term enrichment of secreted proteins down-regulated in shArntl2 cells.
C. Gene expression difference between 482N1 Arntl2 knock-down and shControl cells plotted against gene expression difference between metastasis-derived and TnonMet-derived cell lines. Red dots are genes that encode secreted proteins. Blue dot is Arntl2 itself. Several top secreted factors are labeled.
D, E. Colony formation by 482N1shArntl2 cells under low density plating conditions, quantified in (E). Upper scale bars = 2 cm. Lower scale bars = 1.5 mm. ** p value <0.01; *** p value <0.001.
F. Colony formation by H1792 shControl or shARNTL2 cells under low-density plating conditions, with or without conditioned media (CM) from control cells. ** p value <0.01; *** p value <0.001.
G. Colony formation by 482N1shArntl2#1 cells in low density plating conditions with or without boiled conditioned media. * p value <0.05; ** p value <0.01.
H. Colony formation by 482N1 shArntl2#1 cells under low-density plating conditions with fractionated conditioned media (CM). * p value <0.05; ** p value <0.01.
Panels with error bars show the mean +/− SD. See also Figure S6 and Table S2.
The pro-metastatic function of Arntl2 is conferred by secreted proteins
To test whether these Arntl2-regulated secreted factors drive clonal growth, we determined whether conditioned media from 482N1 cells could rescue the growth defect of 482N1shArntl2 cells plated in low-density conditions. Addition of conditioned media from 482N1shControl cells at the time of plating almost completely rescued the clonogenic ability of 482N1shArntl2 cells (Figure 6D and 6E).
To determine whether human lung adenocarcinoma cells lose clonal growth ability after ARNTL2 knock-down by a similar mechanism, we tested whether conditioned media for human lung adenocarcinoma cell lines also possess soluble factors that promote colony growth in cell culture. The effect of ARNTL2 knock-down on the clonal growth ability of both human lung adenocarcinoma cell lines was rescuable by the addition of conditioned media from their corresponding control cell lines (Figure 6F and data not shown).
Conditioned media could increase the probability of clonal growth due to the presence of either secreted proteins or small molecule metabolites. To determine whether secreted proteins drive the phenotype, we assessed the effect of boiling the conditioned media on its ability to promote clonal growth. Boiling denatures most proteins without affecting small molecules, and boiled conditioned media had almost no ability to increase clonal growth (Figure 6G). We also fractionated the conditioned media and found that soluble factors greater than 3 kDa in size are responsible for the pro-clonogenic activity of the conditioned media (Figure 6H).
Smoc2 is a required component of the pro-metastatic secretome and promotes clonal growth and resistance to anoikis
To identify secreted factors that regulate metastatic ability, we focused on poorly characterized secreted factors that are highly expressed in metastasis-derived cell lines and robustly down-regulated in Arntl2 knock-down cells (Figure 6C). Interestingly, the transcript levels of the extracellular matrix-associated protein Smoc2 was dramatically down-regulated (> 10-fold reduced) in shArntl2 cells and highly expressed (almost 10-fold higher) in metastasis (Met)-derived cell lines (Figure 6C). Smoc2 is a member of the SPARC family of matricellular proteins. Members of this protein family function as non-structural links between the extracellular matrix and cells, modulating, among others, focal adhesions and actin stress fiber organization via activating cellular integrins (De et al., 2003; Sage et al., 1989). Smoc2 is widely expressed during embryonic development, enhances responses to angiogenic growth factors, and mediates cell adhesion(Maier et al., 2008; Rocnik et al., 2006). Smoc2 also marks intestinal stem cells, and its induction in colorectal cancer cells was shown to promote epithelial-mesenchymal transition and confer a stem cell-like phenotype (Shvab et al., 2015).
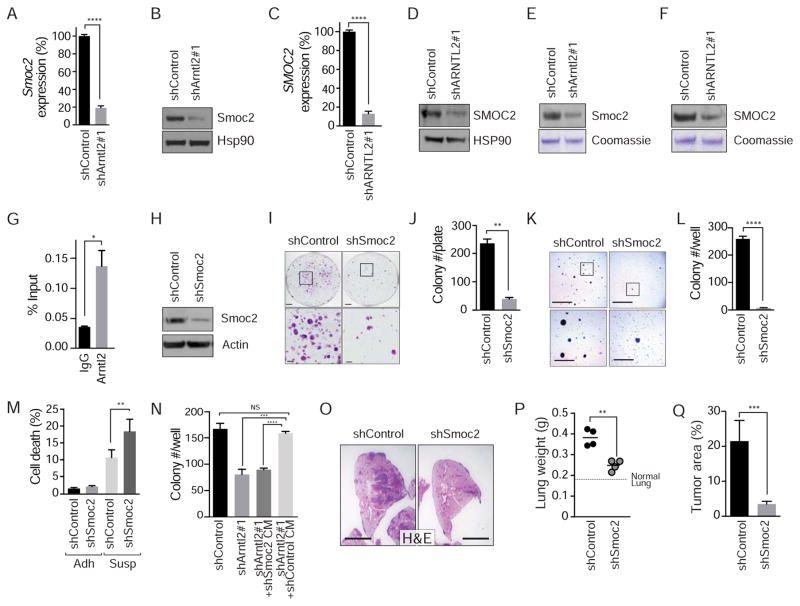
We confirmed reduced Smoc2 in murine 482N1shArntl2 and 889shArntl2 cells, as well as in human H1792shARNTL2 cells (Figure 7A–7D and S7A–S7D). Secreted Smoc2 protein was detectable in conditioned media and consistently reduced upon Arntl2 knock-down (Figure 7E and 7F). We could also detect binding of Arntl2 to the proximal promoter of Smoc2 by ChIP-qPCR, suggesting that Arntl2 may directly regulate expression of Smoc2 (Figure 7G, S7E, and data not shown).
Figure 7. Arntl2-controlled expression of Smoc2 drives clonal growth and metastatic ability.
A, B. Smoc2 expression at the RNA (A) and protein (B) level in 482N1 cells with or without Arntl2 knock-down. Gene expression normalized to Gapdh and shControl. Hsp90 shows loading. **** p value <0.0001.
C. Western blot for Smoc2 protein levels in conditioned media (CM) derived from 482N1 cells with or without Arntl2 knock-down. Coomassie staining shows loading.
D, E. Smoc2 expression at the RNA (D) and protein (E) level in H1792 cells with or without ARNTL2 knock-down. Gene expression normalized to ACTIN and shControl. HSP90 shows loading. **** p value <0.0001.
F. Western blot for SMOC2 protein levels in conditioned media (CM) derived from H1792 cells with or without ARNTL2 knock-down. Coomassie staining shows loading.
G. Chromatin immunoprecipitation of Arntl2 at the Smoc2 proximal promoter (Tile 11) in 482N1 lung adenocarcinoma cells. Data are normalized to input.
H. Western blot for Smoc2 in the 482N1 cell line, with or without Smoc2 knock-down. Actin shows loading.
I, J. 482N1 lung adenocarcinoma colony formation under low-density plating conditions (3x103 cells/10 cm plate) with or without Smoc2 knock-down. Upper scale bars = 2 cm, lower scale bars = 5 mm. Quantified in (J). ** p value <0.01.
K, L. 482N1 lung adenocarcinoma cell growth in anchorage-independent conditions with or without Smoc2 knock-down. Upper scale bars = 5 mm, lower scale bars = 1.5 mm. Quantified in (L). **** p value <0.0001.
M. Quantification of percent DAPIpositive dead cells after plating in adherent (Adh) and suspension (Susp) conditions of 482N1 cells with or without Smoc2 knock-down. *** p value <0.001; * p value <0.05.
N. Colony formation of 482N1shArntl2 cells under low-density plating conditions (1x103 cells/6 well) with or without shSmoc2 conditioned media (CM). Upper scale bars = 5 mm, lower scale bars = 1.5 mm.
O. Representative histology of metastatic lung lesions three weeks after intravenous transplantation of 482N1 cells. Scale bar = 3 mm.
P. Total lung weight per mouse, measured three weeks after intravenous transplant of 482N1 cells, with or without Smoc2 knock-down. Each dot represents a mouse and the bar is the mean. Normal lung weight is indicated. ** p value <0.01. Q. Quantification of tumor area relative to total lung area. *** p value <0.001.
Panels with error bars show the mean +/− SD. See also Figure S7.
To determine the function of Smoc2, we knocked-down Smoc2 in 482N1 cells with two independent shRNAs and assessed cellular phenotypes in cell culture and metastatic ability in vivo (Figure 7H, S7F, and S7G). Smoc2 knock-down did not affect proliferation or cell death under standard culture conditions (Figure S7H–S7J), but clonal growth ability in low-density plating and anchorage-independent growth conditions required Smoc2 (Figure 7I–7L and S7K). Smoc2 knock-down cells also showed increased cell death when plated in suspension growth conditions (Figure 7M and S7L). Consistent with the impaired clonal growth ability of shSmoc2 cells, conditioned media from shSmoc2 cells failed to rescue clonal growth of shArntl2 cells (Figure 7N and S7M). Importantly, Smoc2 was also required for efficient metastatic seeding in vivo, suggesting that Smoc2 is a pro-metastatic factor (Figure 7O–7Q). These results are consistent with Smoc2 being a key Arntl2-regulated pro-metastatic factor.
Arntl2 promotes the expression of pro-metastatic secreted factors in association with Clock
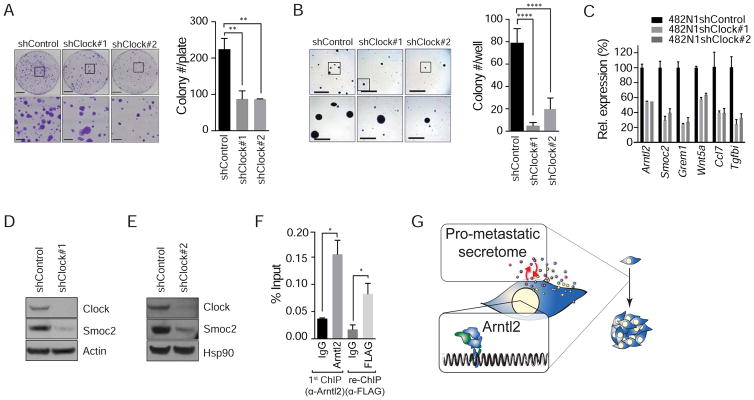
Arntl2 is a bHLH transcription factor that has been shown to interact with several transcriptional partners, including Hif1α and the core circadian rhythm transcription factor Clock (Hogenesch et al., 2000; Sasaki et al., 2009; Shi et al., 2010). We investigated Clock as a potential partner for Arntl2 in driving malignant phenotypes and the expression of pro-metastatic secreted factors. To determine whether lung adenocarcinoma clonal growth is controlled in a Clock-dependent manner, we knocked-down Clock. Stable knock-down of Clock did not alter proliferation or cell death under standard culture conditions (Figure S8A, and S8B and data not shown). Quantification of growth in low-density plating and anchorage-independent conditions indicated that Clock knock-down phenocopied Arntl2 knock-down, with shClock cells forming significantly fewer colonies (Figure 8A–B).
Figure 8. Smoc2 expression is regulated by Arntl2, in association with Clock.
A. 482N1 colony formation under low-density plating conditions (3x103 cells/10 cm plate) with or without Clock knock-down. Upper scale bars = 2 cm, lower scale bars = 5 mm. ** p value <0.01.
B. 482N1 colony formation in anchorage-independent conditions with or without Clock knock-down. Upper scale bars = 5 mm, lower scale bars = 1.5 mm. **** p value <0.0001.
C. qPCR for expression of several Arntl2-regulated genes in Clock knock-down lung adenocarcinoma cells. All gene expression levels are significant at p value < 0.0001 relative to shControl and Gapdh.
D, E. Western blot for Smoc2 protein expression in 482N1 Clock knock-down cell lines. Actin (D) or Hsp90 (E) show loading.
F. Chromatin immunoprecipitation of Arntl2 at the Smoc2 proximal promoter (Tile 11), followed by re-ChIP for 3xFLAG-Clock in 482N1 cells overexpressing 3xFLAG-Clock. * p value < 0.05.
G. Model in which high levels of Arntl2 drive the expression of a pro-metastatic secretome which increases the metastatic ability of single lung adenocarcinoma cells within a foreign environment.
Panels with error bars show the mean +/− SD. See also Figure S8.
Clock was required for the expression of several Arntl2-regulated secreted factors, indicating that Clock may be broadly required for the expression of Arntl2-regulated secreted factors (Figure 8C). In particular, Clock knock-down robustly reduced Smoc2 mRNA and protein (Figure 8C–E). To determine whether Arntl2 and Clock co-bind the Smoc2 proximal promoter we performed ChIP-reChIP on lung cancer cells engineered to express FLAG-tagged Clock (Figure S8C). Immunoprecipitation of Arntl2 followed by immunoprecipitation with either an anti-FLAG or anti-Clock antibody showed enrichment for co-occupancy at the Smoc2 promoter (Figure 8F and S8D).
In addition to forming a transcription complex with Arntl2, Clock (in a complex with Arntl1/Bmal1) can regulate Arntl2 expression in some cell types (Bunger et al., 2000; Shi et al., 2010). In shClock cells, Arntl2 mRNA was reduced, resulting in a modest reduction of Arntl2 protein (Figure S8E). Expression of Arntl2 cDNA in shClock cells restored Arntl2 protein level back to the level in shControl cells, but was not sufficient to restore the expression of Smoc2 and other secreted factors (Figure S8E and S8F). Collectively, these data suggest a model in which Arntl2 and Clock control the expression of pro-metastatic secreted factors to enable metastatic self-sufficiency and the expansion of single cancer cells into metastases (Figure 8G).
DISCUSSION
We integrated gene expression analyses of murine and human lung adenocarcinoma with functional studies in cell culture and in vivo to identify the transcription factor Arntl2 as a key driver of lung adenocarcinoma metastatic self-sufficiency. Our data support a model in which a subset of human lung adenocarcinomas progress to a less differentiated Arntl2-positive state. Arntl2 itself is not required to maintain the poorly differentiated state, but rather cooperates with Clock to drive the expression of a complex pro-metastatic secretome that facilitates metastatic seeding and outgrowth (Figure 8G). Our unbiased analyses highlighted Smoc2 as a potential regulator of metastatic ability, and functional assays suggest that Smoc2 is a pro-metastatic autocrine matricellular protein.
Lung adenocarcinoma cell growth in vitro and as a subcutaneous tumor mass was not affected by knock-down of Arntl2. In these conditions, individual cancer cells are in close proximity to one another and gain little, if any, advantage from Arntl2 expression and the secreted factors that Arntl2 controls. Thus, cancer cells in these conditions may not require Arntl2-controlled factors because they either cooperate to establish high enough concentrations of these or other secreted factors, or because other cell-cell contacts abrogate the need for these pro-clonal growth factors. Foreign organ environments could also impose additional inhibitory signals that are incidentally anti-metastatic, but can be overcome by the signals induced by the Arntl2-regulated secreted factors.
How do Arntl2 and Smoc2 promote metastasis? Arntl2 and Smoc2 knock-down increase anoikis, suggesting that part of the pro-metastatic function of these factors might be to limit cell death during the metastatic process. While most circulating and disseminated cancer cells seem to be single cells, a reduced requirement for metastatic self-sufficiency could be one factor that increases the metastatic ability of clusters of cancer cells (Aceto et al., 2014). While Smoc2 has not been well-studied in any disease context, it is a marker of colon stem cells, it modulates the mitogenic and angiogenic effects of several growth factors, and regulates myeloid development in zebrafish (Mommaerts et al., 2014; Rocnik et al., 2006; Shvab et al., 2015). Smoc2 has been shown to regulate keratinocyte adhesion through integrin binding. Integrins are established regulators of anoikis resistance, tissue stem cell maintenance, and cancer stem cell phenotypes (Desgrosellier and Cheresh, 2010; Prowse et al., 2011). Our cell culture and in vivo data implicating Smoc2 function in metastasis suggest that the ability of Smoc2 to link structural extracellular matrix proteins to cancer cell integrins may provide critical molecular signals for survival, cell cycle progression, and other cancer stem cell properties during lung adenocarcinoma metastasis.
In addition to Smoc2, many other secreted factors that have been previously linked to metastases are down-regulated in Arntl2 knock-down cells and are more highly expressed in lung adenocarcinoma metastases from the autochthonous mouse model. In particular Wnt5a, Il11, and Cxcl5 have been implicated as pro-metastatic factors in other cancer types (Bo et al., 2013; Hanaki et al., 2012; Kang et al., 2013; Weeraratna et al., 2002; Zhou et al., 2014). Additional work will be required to determine which other Arntl2-regulated secreted factors influence additional aspects of lung cancer progression. While some of these factors may cooperate with Smoc2 in an autocrine manner, others may also impact stromal cells within the primary tumor or metastatic site.
A relationship between clonal growth ability during metastasis and stem cell characteristics seems to be a tautology, but this is perhaps only true in that cancer cells that seed metastases are obligated to have these stem-like qualities. Conversely, cancer cells that sustain primary tumor growth may be inefficient at seeding metastases, as they may remain dependent on signals from the established tumor environment and therefore lack self-sufficient growth ability. Normal tissue stem cells lose their stemness when removed from their niche, suggesting that some of the factors regulated by Arntl2 increase the ability of lung cancer cells to maintain their tumor-initiating characteristics.
Given the critical role of ARNTL2 in driving metastatic ability of lung adenocarcinoma, it will also be of interest to understand what leads to ARNTL2 up-regulation. The lung differentiation transcription factor Nkx2-1 suppresses lung adenocarcinoma metastatic ability, but Nkx2-1 re-expression in Nkx2-1negative lung adenocarcinoma cells was not sufficient to repress Arntl2 (Figure S2F, S2G and data not shown). Nkx2-1 knock-down also does not lead to Arntl2 expression, and not all poorly differentiated human or mouse lung adenocarcinomas express ARNTL2 at a high level, suggesting that loss of differentiation alone is insufficient to induce its expression. Arntl2 expression could be regulated entirely independent of Nkx2-1, or the alterations that lead to Nkx2-1 down-regulation could reciprocally up-regulate Arntl2 expression.
How different genes and pathways interact to enable metastatic ability is also an area of great importance. While Nkx2-1 and Arnlt2 do not regulate the expression of one another, gain- and loss-of-function experiments in metastatic and non-metastatic cell lines can suggest how they functionally interact. In non-metastatic Nkx2-1positiveArntl2negative cells, Nkx2-1 knock-down is sufficient to increase phenotypes associated with metastatic ability while Arntl2 expression is not (Winslow et al., 2011 and Figure 4H). Conversely, in Nkx2-1negativeArntl2positive metastatic cancer cells either Nkx2-1 expression or Arntl2 knock-down is sufficient to reduce metastatic ability. This data is consistent with a model in which Nkx2-1-mediated differentiation is dominant over Arntl2’s ability to promote phenotypes associated with metastatic ability. Thus, the pro-metastatic effect of Arntl2 is strongest in cancer cells that have transitioned to a poorly differentiated state.
By comparing ARNTL2 expression with patient outcome across 20 other cancer types, we found that high ARNTL2 is also a poor prognostic marker in low-grade glioma, renal clear cell carcinoma, and, to a lesser extent, pancreatic adenocarcinoma (Figure S8G–J). While there is no obvious relationship between these cancers at the genomic, developmental, or pathophysiological level, we speculate that the cellular phenotypes conferred by high ARNTL2 expression may promote malignant growth or metastatic ability in a subset of patients with these other diverse cancer types.
The ability of single cancer cells to establish inoperable metastases in secondary organs is a major impediment to successful therapy. The identification of a pro-metastatic secretome controlled by a single transcription factor provides the potential for targeting the metastatic self-sufficiency of these cells in an effort to curb the metastatic process prior to expansion at a foreign site. Our findings illustrate how cancer cells can transcriptionally prepare to survive as single entities through the challenges of the metastatic process and in hostile environments prior to clonogenic expansion.
EXPERIMENTAL PROCEDURES
Mouse and human lung adenocarcinoma gene expression analysis
Expression of Arntl2 in TnonMet, TMet, and Met KP murine cell lines is from a previously generated dataset (Winslow et al., 2011). For the analysis of Arntl2 expression in ex vivo mouse lung adenocarcinoma samples, we used Fluidigm qPCR to assess Arntl2 expression as previously described (Caswell et al., 2014). Human lung adenocarcinoma gene expression and clinical data for lung adenocarcinoma was from TCGA Data Portal. Data was z score normalized. For each profiled gene (with non-zero expression in at least one sample and sufficient standard deviation), we tested 7 z score thresholds (−1, −0.5, −0.25,0,0.25,0.5,1) and determined the difference in Kaplan-Meier survival curves for individuals with expression levels below and above the chosen z score threshold. The threshold that led to the minimum p value across 10-fold cross-validation is indicated. From this threshold, we calculated the q value and the hazard ratio between the two groups.
Histology and RNA in situ hybridization
Lung and liver samples were fixed in 4% formalin and paraffin embedded. Hematoxylin and eosin staining was performed using standard methods. Percent tumor area was calculated using ImageJ. ARNLT2 mRNA expression was assessed in human lung adenocarcinoma tissue microarrays (LC20813 and LC20815; US Biomax) with a custom RNAscope probe for ARNTL2 (Advanced Cell Diagnostics). RNA hybridization and development was performed according to manufacturer’s instructions using the RNAscope 2.0 HD Detection Kit (RED) using an Advanced Cell Diagnostics HybEZ™ Oven. ARNTL2 signal was scored as positive for red signal dots with high signal-to-background ratio, and crisp, clear edges. Only adenocarcinoma cores were included in our analysis. Cancer cell cytological differentiation was provided by the supplier.
Cell lines and lentiviral gene knock-down
Mouse Arntl2, Smoc2, and Clock and human ARNTL2 were knocked down using pLKO/Puro lentiviral vectors. Transduced cells were selected with puromycin and knockdown was confirmed by qPCR and western blotting. 482N1 cells have been described (Winslow et al., 2011). 889DTC cells were generated from disseminated cancer cells from the pleural cavity of a KPT mouse. H1792 and H2009 were from ATCC.
Mouse intravenous and intrasplenic transplantation
For intravenous transplantation 105 482N1 cells in 200 μL PBS were injected into the lateral tail vein of male 129/Bl6 F1 mice (Jackson Laboratories, Stock number 101043). For intrasplenic injection 105 482N1 cells in 50 μL PBS were injected into male 129/Bl6 F1 mice using standard methods (Li et al., 2013). For subcutaneous injections 5x104 889DTC cells in 50 μL PBS were injected into each flank and shoulder of immune compromised NOD/Scid/γC (NSG) mice (The Jackson Laboratories, Stock number 005557). Intravenously and intrasplenically injected mice were analyzed 21 days after transplantation. All experiments were performed in accordance with Stanford University Animal Care and Use Committee guidelines.
RNA-seq library preparation and analysis
RNA-sequencing libraries were prepared using the Illumina TruSeq v2 kit according to manufacturer’s instructions. Total RNA was isolated from each cell line (482N1shControl, 482N1shArntl2#1, and 482N1shArntl2#2) in duplicate using the Qiagen RNAeasy mini kit. For each sample, 0.5 μg of total RNA was used for library construction. High-throughput sequencing was performed an Illumina HiSeq 2000 for 100 bp paired-end reads. See Supplemental Experimental Procedures for RNA-seq data analysis.
Cell culture and conditioned media assays
Low-density plating, anchorage independent growth, proliferation, migration, anoikis, and cell death assays were performed using standard methods (see Supplemental Experimental Procedures). Colony number was assessed using ImageJ. Conditioned media was generated by seeding 5 x 106 mouse or human shControl cells per 15-cm dish (or 1x106 per 10-cm dish) for 24 hours. Once confluency was reached, media was replaced and 24 hours later, the resulting conditioned media filtered through a 0.45-μm filter, and applied at ¼ dilution into complete growth media. For heat-inactivation, filtered media was boiled for 10 min. Fractionation was done using Macrosep Advance Devices with a MWCO of 3 kDa (Pall Corporation). The retentate and eluate were collected separately and resuspended to the original pre-fractionated volume with fresh DMEM.
Chromatin Immunoprecipitation followed by reChIP/qPCR
Cells were fixed in 1% formaldehyde for 10 min at room temperature, followed by quenching with glycine, and snap frozen into aliquots of 2x106 cells. Fixed cells were stored in liquid nitrogen until use. Cell pellets were resuspended in 130 μL TE and sonicated using a Covaris S220 sonicator for 5 cycles of 50 s each, with 5% duty factor, and 200 cycles per burst. Chromatin immunoprecipitation and reChIP was done according to the Rockland-Inc ChIP protocol at www.rockland-inc.com. See Supplemental Experimental Procedures for additional details.
Supplementary Material
SIGNIFICANCE.
Most cancer patients die due to the growth of metastases and metastatic lung cancer is a leading cause of cancer deaths. While cancer cells within primary tumors may benefit from neighboring cancer cells, metastatic cells must possess the ability to expand from a single cell in a foreign environment. Uncovering the mechanisms by which cancer cells acquire this metastatic self-sufficiency could provide avenues for therapeutic intervention and represent clinical biomarkers of aggressive disease. We identify a pro-metastatic secretome, induced by the transcription factor Arntl2, that enables single cancer cells to expand into tissue-destructive metastases, thus providing a molecular understanding of how a subset of metastatic lung cancer cells gain metastatic self-sufficiency.
HIGHLIGHTS.
High ARNTL2 is a top predictor of lung adenocarcinoma patient outcome
Arntl2 is required for metastatic ability in vivo and clonal growth in cell culture
An Arntl2-driven pro-metastatic secretome controls metastatic self-sufficiency
Smoc2 is an Arntl2/Clock-dependent pro-metastatic secreted factor
Acknowledgments
We thank Pauline Chu, Sopheak Sim, Aidan Winters, and Johanna Huelsmann for technical assistance; Leanne Sayles for reagents; Matthew Fish for guidance on in situ hybridization; Sean Dolan and Alexandra Orantes for administrative support; David Feldser and members of the Winslow laboratory for helpful comments. We thank the Stanford PAN Facility and Stanford Shared FACS Facility. This work was supported by NIH R01-CA175336 (to M.M.W.), an American Lung Association Biomedical Research Grant (to M.M.W.), and in part by the Stanford Cancer Institute support grant (NIH P30-CA124435). J.J.B. was supported by NIH F32-CA189659. C-H.C. was funded by a Stanford Dean’s Fellowship and an American Lung Association Fellowship. P.G.G. was supported by the Stanford Biomedical Informatics Training Grant from the National Library of Medicine (LM-07033) and a Bio-X Stanford Interdisciplinary Graduate Fellowship. Z.N.R. was supported by a Stanford Graduate Fellowship and a National Science Foundation Predoctoral fellowship. C.W.M. is an Anne T. and Robert M. Bass Stanford Graduate Fellow. D.R.C. was supported by NIH T32CA09302 and a National Science Foundation Predoctoral fellowship. A.K. is supported by an Alfred Sloan Fellowship. U.H. was supported by grants from the Deutsche Forschungsgemeinschaft (HA 2263/2) and from the Köln Fortune Program of the Medical Faculty of the University of Cologne. E.A.S.C. is supported by NIH R01-CA157510.
Footnotes
ACCESSION NUMBERS
RNA-seq data on 482N1shControl and 482N1shArntl2 cell lines have been deposited to GEO under accession number GSE72942.
Supplemental Information includes 8 figures and 2 tables.
The authors declare no conflicts of interest.
AUTHOR CONTRIBUTIONS
J.J.B. and M.M.W. designed the study, and analyzed and interpreted the data; J.J.B., C-H.C., D.R.C., and M.M.W. performed the experiments; P.G.G. and Z.N.R. analyzed human TCGA data under supervision of A.K., C.W.M. analyzed isoform data, E.A.S.C. and U.H. provided reagents; A.J.C. provided pathology expertise; J.J.B. and M.M.W. wrote the manuscript with comments and feedback from all authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA, Yu M, Pely A, Engstrom A, Zhu H, et al. Circulating Tumor Cell Clusters Are Oligoclonal Precursors of Breast Cancer Metastasis. Cell. 2014;158:1110–1122. doi: 10.1016/j.cell.2014.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bafico A, Liu G, Goldin L, Harris V, Aaronson SA. An autocrine mechanism for constitutive Wnt pathway activation in human cancer cells. Cancer Cell. 2004;6:497–506. doi: 10.1016/j.ccr.2004.09.032. [DOI] [PubMed] [Google Scholar]
- Blanco MA, LeRoy G, Khan Z, Ale3kovi3 M, Zee BM, Garcia BA, Kang Y. Global secretome analysis identifies novel mediators of bone metastasis. Cell Res. 2012;22:1339–1355. doi: 10.1038/cr.2012.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bo H, Zhang S, Gao L, Chen Y, Zhang J, Chang X, Zhu M. Upregulation of Wnt5a promotes epithelial-to-mesenchymal transition and metastasis of pancreatic cancer cells. BMC Cancer. 2013;13:496. doi: 10.1186/1471-2407-13-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caswell DR, Chuang CH, Yang D, Chiou SH, Cheemalavagu S, Kim-Kiselak C, Connolly A, Winslow MM. Obligate Progression Precedes Lung Adenocarcinoma Dissemination. Cancer Discov. 2014;4:781–789. doi: 10.1158/2159-8290.CD-13-0862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung WKC, Nguyen DX. Lineage factors and differentiation states in lung cancer progression. Oncogene. 2015 Mar 30; doi: 10.1038/onc.2015.85. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung WKC, Zhao M, Liu Z, Stevens LE, Cao PD, Fang JE, Westbrook TF, Nguyen DX. Control of alveolar differentiation by the lineage transcription factors GATA6 and HOPX inhibits lung adenocarcinoma metastasis. Cancer Cell. 2013;23:725–738. doi: 10.1016/j.ccr.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox TR, Rumney RMH, Schoof EM, Perryman L, Høye AM, Agrawal A, Bird D, Latif NA, Forrest H, Evans HR, et al. The hypoxic cancer secretome induces pre-metastatic bone lesions through lysyl oxidase. Nature. 2015;522:106–110. doi: 10.1038/nature14492. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- De S, Chen J, Narizhneva NV, Heston W, Brainard J, Sage EH, Byzova TV. Molecular pathway for cancer metastasis to bone. J Biol Chem. 2003;278:39044–39050. doi: 10.1074/jbc.M304494200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desgrosellier JS, Cheresh DA. Integrins in cancer: biological implications and therapeutic opportunities. Nat Rev Cancer. 2010;10:9–22. doi: 10.1038/nrc2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ell B, Kang Y. Transcriptional control of cancer metastasis. Trends Cell Biol. 2013;23:603–611. doi: 10.1016/j.tcb.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidler IJ. The pathogenesis of cancer metastasis: the “seed and soil” hypothesis revisited. Nat Rev Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- Gy3rffy B, Surowiak P, Budczies J, Lánczky A. Online Survival Analysis Software to Assess the Prognostic Value of Biomarkers Using Transcriptomic Data in Non-Small-Cell Lung Cancer. PLoS ONE. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaki H, Yamamoto H, Sakane H, Matsumoto S, Ohdan H, Sato A, Kikuchi A. An Anti-Wnt5a Antibody Suppresses Metastasis of Gastric Cancer Cells In Vivo by Inhibiting Receptor-Mediated Endocytosis. Mol Cancer Ther. 2012;11:298–307. doi: 10.1158/1535-7163.MCT-11-0682. [DOI] [PubMed] [Google Scholar]
- Hogenesch JB, Gu Y-Z, Moran SM, Shimomura K, Radcliffe LA, Takahashi JS, Bradfield CA. The Basic Helix-Loop-Helix-PAS Protein MOP9 Is a Brain-Specific Heterodimeric Partner of Circadian and Hypoxia Factors. J Neurosci. 2000;20:RC83–RC83. doi: 10.1523/JNEUROSCI.20-13-j0002.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, Jacks T. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- Kang Y, Siegel PM, Shu W, Drobnjak M, Kakonen SM, Cordón-Cardo C, Guise TA, Massagué J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- Kimura S, Hara Y, Pineau T, Fernandez-Salguero P, Fox CH, Ward JM, Gonzalez FJ. The T/ebp null mouse: thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Genes Dev. 1996;10:60–69. doi: 10.1101/gad.10.1.60. [DOI] [PubMed] [Google Scholar]
- Korpal M, Ell BJ, Buffa FM, Ibrahim T, Blanco MA, Celià-Terrassa T, Mercatali L, Khan Z, Goodarzi H, Hua Y, et al. Direct targeting of Sec23a by miR-200s influences cancer cell secretome and promotes metastatic colonization. Nat Med. 2011;17:1101–1108. doi: 10.1038/nm.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CMC, Gocheva V, Oudin MJ, Bhutkar A, Wang SY, Date SR, Ng SR, Whittaker CA, Bronson RT, Snyder EL, Gertler FB. Foxa2 and Cdx2 cooperate with Nkx2-1 to inhibit lung adenocarcinoma metastasis. Genes Dev. 2015;29:1850–1862. doi: 10.1101/gad.267393.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CMC, Chen G, Dayton TL, Kim-Kiselak C, Hoersch S, Whittaker CA, Bronson RT, Beer DG, Winslow MM, Jacks T. Differential Tks5 isoform expression contributes to metastatic invasion of lung adenocarcinoma. Genes Dev. 2013;27:1557–1567. doi: 10.1101/gad.222745.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Xu Z, Du W, Zhang Z, Wei Y, Wang H, Zhu Z, Qin L, Wang L, Niu Q, et al. Aiolos promotes anchorage independence by silencing p66Shc transcription in cancer cells. Cancer Cell. 2014;25:575–589. doi: 10.1016/j.ccr.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Ghosh S, Wang Z, Hunter T. Downregulation of caveolin-1 function by EGF leads to the loss of E-cadherin, increased transcriptional activity of beta-catenin, and enhanced tumor cell invasion. Cancer Cell. 2003;4:499–515. doi: 10.1016/s1535-6108(03)00304-0. [DOI] [PubMed] [Google Scholar]
- Maier S, Paulsson M, Hartmann U. The widely expressed extracellular matrix protein SMOC-2 promotes keratinocyte attachment and migration. Exp Cell Res. 2008;314:2477–2487. doi: 10.1016/j.yexcr.2008.05.020. [DOI] [PubMed] [Google Scholar]
- Mazzoccoli G, Pazienza V, Panza A, Valvano MR, Benegiamo G, Vinciguerra M, Andriulli A, Piepoli A. ARNTL2 and SERPINE1: potential biomarkers for tumor aggressiveness in colorectal cancer. J Cancer Res Clin Oncol. 2012;138:501–511. doi: 10.1007/s00432-011-1126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAllister SS, Weinberg RA. The tumour-induced systemic environment as a critical regulator of cancer progression and metastasis. Nat Cell Biol. 2014;16:717–727. doi: 10.1038/ncb3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo W, Chen J, Patel A, Zhang L, Chau V, Li Y, Cho W, Lim K, Xu J, Lazar AJ, et al. CXCR4/CXCL12 Mediate Autocrine Cell- Cycle Progression in NF1-Associated Malignant Peripheral Nerve Sheath Tumors. Cell. 2013;152:1077–1090. doi: 10.1016/j.cell.2013.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mommaerts H, Esguerra CV, Hartmann U, Luyten FP, Tylzanowski P. Smoc2 modulates embryonic myelopoiesis during zebrafish development. Dev Dyn. 2014;243:1375–1390. doi: 10.1002/dvdy.24164. [DOI] [PubMed] [Google Scholar]
- Mu D. The complexity of thyroid transcription factor 1 with both pro- and anti-oncogenic activities. J Biol Chem. 2013;288:24992–25000. doi: 10.1074/jbc.R113.491647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy-Ullrich JE, Sage EH. Revisiting the matricellular concept. Matrix Biol J Int Soc Matrix Biol. 2014;37:1–14. doi: 10.1016/j.matbio.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DX, Bos PD, Massagué J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009a;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- Nguyen DX, Chiang AC, Zhang XHF, Kim JY, Kris MG, Ladanyi M, Gerald WL, Massagué J. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell. 2009b;138:51–62. doi: 10.1016/j.cell.2009.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obenauf AC, Zou Y, Ji AL, Vanharanta S, Shu W, Shi H, Kong X, Bosenberg MC, Wiesner T, Rosen N, et al. Therapy-induced tumour secretomes promote resistance and tumour progression. Nature. 2015;520:368–372. doi: 10.1038/nature14336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskarsson T, Acharyya S, Zhang XHF, Vanharanta S, Tavazoie SF, Morris PG, Downey RJ, Manova-Todorova K, Brogi E, Massagué J. Breast cancer cells produce tenascin C as a metastatic niche component to colonize the lungs. Nat Med. 2011;17:867–874. doi: 10.1038/nm.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prowse ABJ, Chong F, Gray PP, Munro TP. Stem cell integrins: implications for ex-vivo culture and cellular therapies. Stem Cell Res. 2011;6:1–12. doi: 10.1016/j.scr.2010.09.005. [DOI] [PubMed] [Google Scholar]
- Rocnik EF, Liu P, Sato K, Walsh K, Vaziri C. The novel SPARC family member SMOC-2 potentiates angiogenic growth factor activity. J Biol Chem. 2006;281:22855–22864. doi: 10.1074/jbc.M513463200. [DOI] [PubMed] [Google Scholar]
- Sage H, Vernon RB, Funk SE, Everitt EA, Angello J. SPARC, a secreted protein associated with cellular proliferation, inhibits cell spreading in vitro and exhibits Ca+2-dependent binding to the extracellular matrix. J Cell Biol. 1989;109:341–356. doi: 10.1083/jcb.109.1.341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M, Yoshitane H, Du NH, Okano T, Fukada Y. Preferential inhibition of BMAL2-CLOCK activity by PER2 reemphasizes its negative role and a positive role of BMAL2 in the circadian transcription. J Biol Chem. 2009;284:25149–25159. doi: 10.1074/jbc.M109.040758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenhard JA, Eren M, Johnson CH, Vaughan DE. Alternative splicing yields novel BMAL2 variants: tissue distribution and functional characterization. Am J Physiol Cell Physiol. 2002;283:C103–C114. doi: 10.1152/ajpcell.00541.2001. [DOI] [PubMed] [Google Scholar]
- Shedden K, Taylor JMG, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, Eschrich S, Jurisica I, Giordano TJ, Misek DE, et al. Gene expression–based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Hida A, McGuinness OP, Wasserman DH, Yamazaki S, Johnson CH. Circadian Clock Gene Bmal1 Is Not Essential After All; Functional Replacement with its Paralog, Bmal2. Curr Biol CB. 2010;20:316–321. doi: 10.1016/j.cub.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shvab A, Haase G, Ben-Shmuel A, Gavert N, Brabletz T, Dedhar S, Ben-Ze’ev A. Induction of the intestinal stem cell signature gene SMOC-2 is required for L1-mediated colon cancer progression. Oncogene 2015. 2015 Apr 27; doi: 10.1038/onc.2015.127. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2015;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeraratna AT, Jiang Y, Hostetter G, Rosenblatt K, Duray P, Bittner M, Trent JM. Wnt5a signaling directly affects cell motility and invasion of metastatic melanoma. Cancer Cell. 2002;1:279–288. doi: 10.1016/s1535-6108(02)00045-4. [DOI] [PubMed] [Google Scholar]
- Wilson TR, Lee DY, Berry L, Shames DS, Settleman J. Neuregulin-1-Mediated Autocrine Signaling Underlies Sensitivity to HER2 Kinase Inhibitors in a Subset of Human Cancers. Cancer Cell. 2011;20:158–172. doi: 10.1016/j.ccr.2011.07.011. [DOI] [PubMed] [Google Scholar]
- Winslow MM, Dayton TL, Verhaak RGW, Kim-Kiselak C, Snyder EL, Feldser DM, Hubbard DD, DuPage MJ, Whittaker CA, Hoersch S, et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011;473:101–104. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou SL, Dai Z, Zhou ZJ, Chen Q, Wang Z, Xiao YS, Hu ZQ, Huang XY, Yang GH, Shi YH, et al. CXCL5 contributes to tumor metastasis and recurrence of intrahepatic cholangiocarcinoma by recruiting infiltrative intratumoral neutrophils. Carcinogenesis. 2014;35:597–605. doi: 10.1093/carcin/bgt397. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Aref AR, Cohoon TJ, Barbie TU, Imamura Y, Yang S, Moody SE, Shen RR, Schinzel AC, Thai TC, et al. Inhibition of KRAS-driven tumorigenicity by interruption of an autocrine cytokine circuit. Cancer Discov. 2014;4:452–465. doi: 10.1158/2159-8290.CD-13-0646. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.