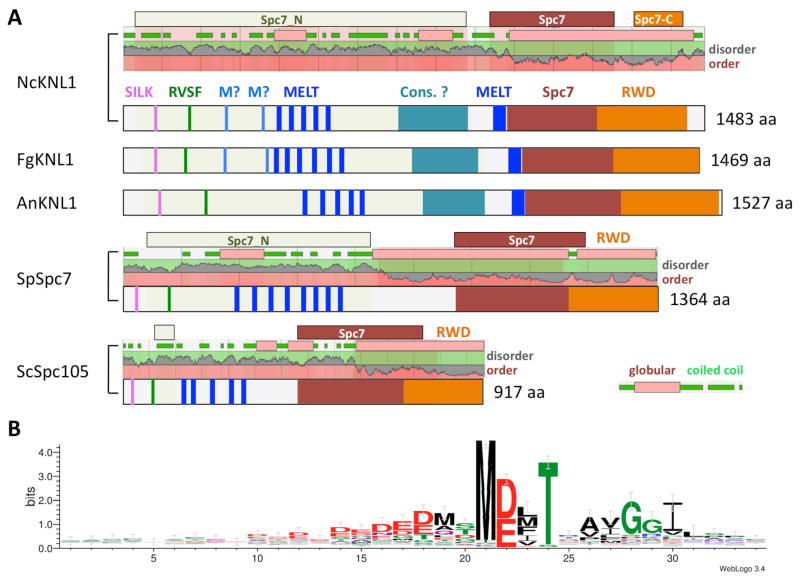
FIG. 5.
Structural features of fungal Knl1/Spc7/Spc105 proteins. A. Cartoons showing important features of Knl1 homologs from fungi. Extent of Pfam domains (Spc7, Spc_N, Spc-C) are shown above plots depicting predicted disorder or order of proteins, and coiled-coil (green) or globular domains (pink). Knl1 homologs from N. crassa (NcKnl1), F. graminearum (FgKnl1) and A. nidulans (AnKnl1) are shown. Overall distribution of hallmark motifs (SILK, RVSF, MELT, Spc7 and tandem RWD domain) is similar in the three examples. The number and type of MELT motifs (MELT and M?) is different, however, and compared to the S. cerevisiae (ScSpc105) and S. pombe (SpSpc7), as well as the metazoan homologs, a potential additional MELT motif immediately before the Spc7 domain was identified. Also unique to filamentous ascomycetes is a conserved globular domain (“Cons.?”) of unknown function. The sequence of this motif is family or order-specific. B. Logo plot of 106 MELT motifs identified in 18 species (Neurospora crassa, Podospora anserina, Magnaporthe oryzae, Fusarium graminearum, Trichoderma reesei, Metarhizium album, Botryotinia fuckeliana, Aspergillus nidulans, Talaromyces marneffei, Arthroderma benhamiae, Histoplasma capsulatum, Coccidioides immitis, Zymoseptoria brevis, Leptosphaeria maculans, Pyronema omphalodens, Candida albicans, Saccharomyces cerevisiae, Schizosaccharomyces pombe) identified M-D/E-L/M/F-T as the preferred motif. Preceding residues are often D/E, the immediately following residues show preference for xAYGGI. Logo was generated by WebLogo 3.4 (Crooks and others, 2004).