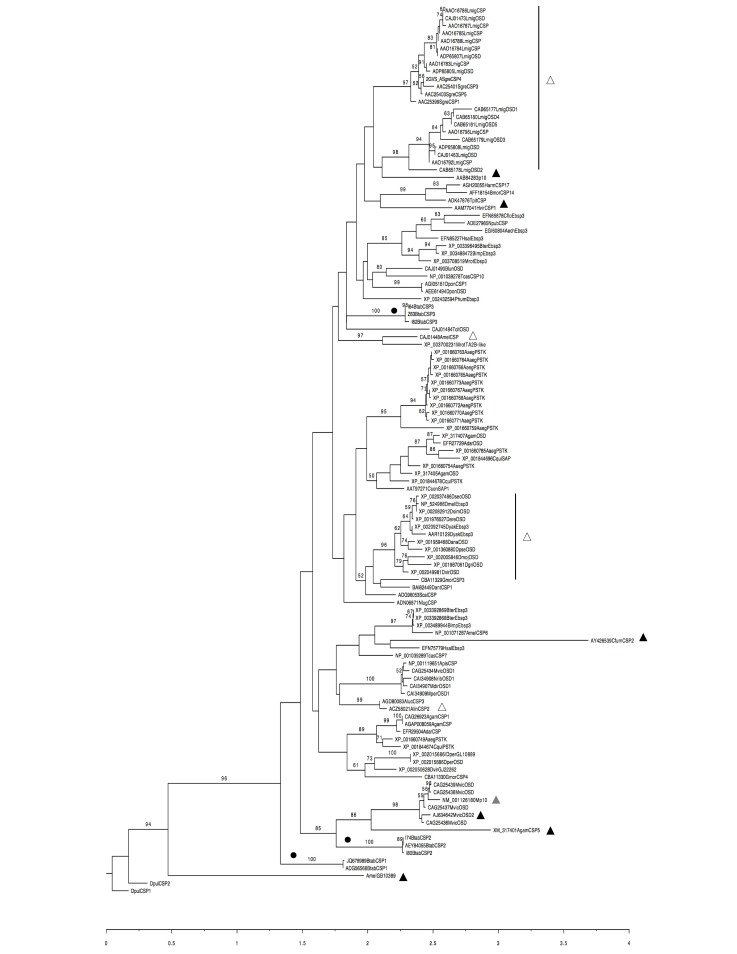
Fig 3. Phylogenetic analysis of amino acid sequences from B. tabaci CSP1, CSP2, CSP3 and orthologs in other insect species (RAxMLGUI).
Aech: Acromyrmex echinatior, Apis: Acyrthosiphon pisum, Alin: Adelphocoris lineolatus, Aaeg: Aedes aegypti, Adar: Anopheles darlingi, Agam: Anophales gambiae, Amel: Apis mellifera, Aluc: Apolygus lucorum, Blun: Biphyllus lunatus, Bter: Bombus terrestris, Bimp: Bombus impatiens, Bmor: Bombyx mori, Cflo: Camponotus floridanus, Cson: Culicoides sonorensis, Dper/Dvir/Dyak: Drosophila persimilis/virilis/yakuba, Dpon: Dendroctonus ponderosae, Gmor: Glossina morsitans morsitans, Hsal: Harpegnatus saltator, Harm: Heliothis armigera, Lmig: Locusta migratoria, Mrot: Megachile rotundata, Mvic: Megoura viciae, Mdir: Metopolophium dirhodum, Mper: Myzus persicae, Npub: Nylanderia pubens, Nlug: Nilaparvata lugens, Nrib: Nasonovia ribis nigri, Phum: Pediculus humanus corporis, Sgre: Schistocerca gregaria, Scal: Stomoxys calcitrans, Tpit: Thaumetopoea pityocampa, Tcit: Toxoptera citricida, Tcas: Tribolium castaneum. Sequences are all published in NCBI. B. tabaci CSP1, CSP2 and CSP3 sequences are from JQ678989-ADG56568, KM078671- KM078679 and KM078694- KM078697, respectively. CSP sequences from Daphnia pulex (DpulCSP1 and DpulCSP2; ABH88167, ABH88166) are used as an outgroup. The triangles indicate the CSP proteins that have been functionally characterized. Black triangles: developmental genes, grey triangles: effector genes, white triangles: olfactory genes.