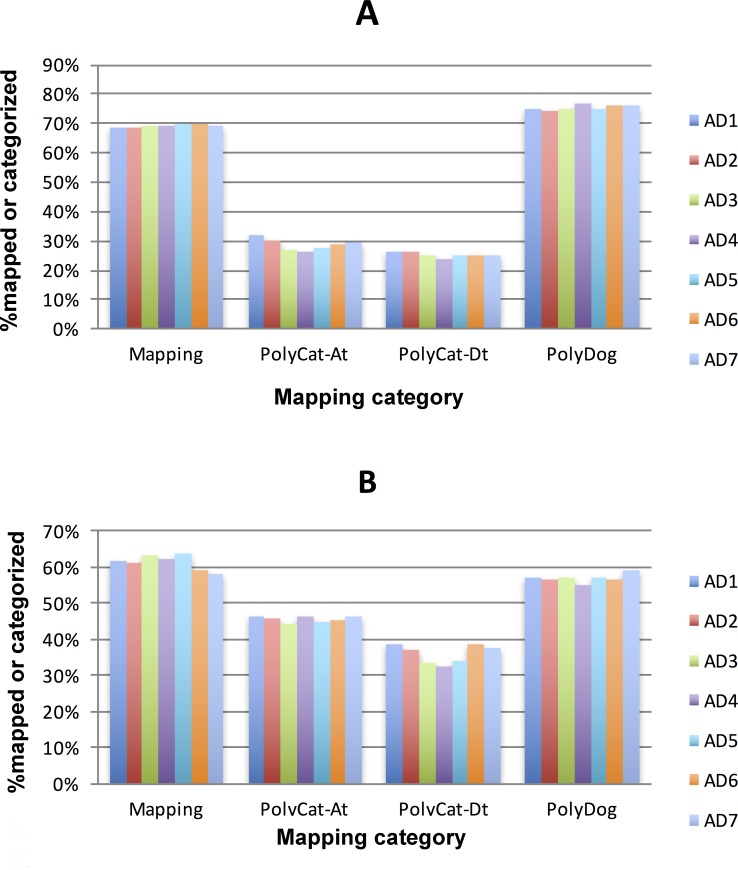
Fig 1. Effectiveness of mapping with GSNAP and categorization with PolyCat and PolyDog.
PolyDog has higher categorization rates because it allows for genome-wide categorization, instead of relying on known homoeo-SNPs in regions present in both genomes. The percentage of trimmed reads successfully mapped from each species (AD1-AD7) to the A2 reference (A) and the D5 reference (B) is shown. For each reference, the percentage of mapped reads from each species (AD1-AD7) categorized to the AT-genome by PolyCat, DT-genome by PolyCat, or to the genome of the reference sequence by PolyDog is also shown.