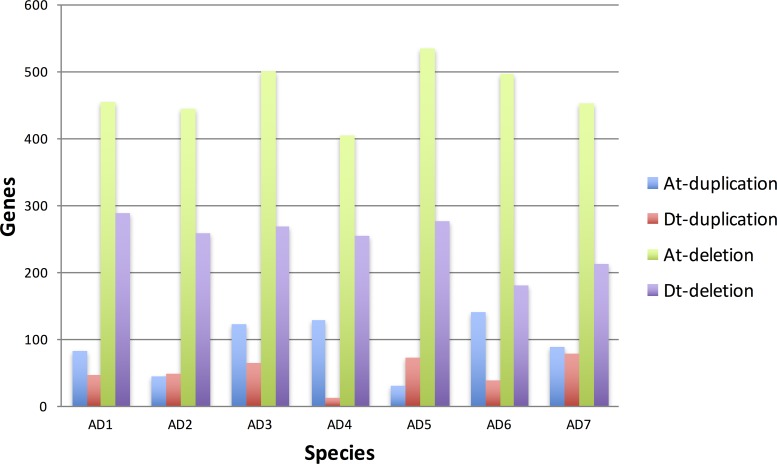
Fig 2. Copy number variants were generally more common in the larger AT-genome than in the smaller DT-genome, and deletions were more common than duplications, in accordance with the idea of reciprocal gene loss.
Duplications and deletions were identified in each genome of each species, relative to the extant diploid relative. CNVkit detected CNVs and determined their sizes using a log base 2 threshold of 1.0 for duplications and -1.0 for deletions. Blue indicates duplications in AT-genomes compared to A2 individuals. CNVkit identified duplications and deletions, with a minimum threshold of 2-fold difference.