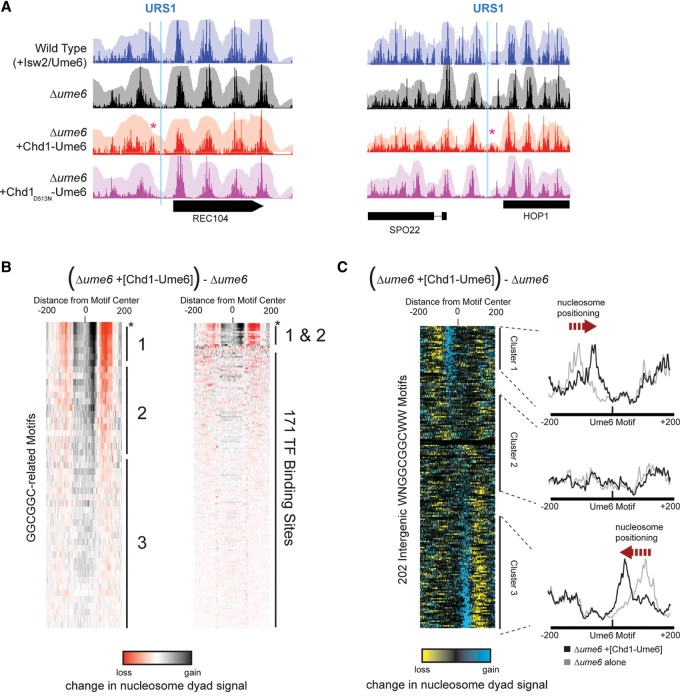
Figure 2.
Chd1-Ume6 organizes chromatin at genomic Ume6 target sequences. (A) MNase-seq signal of nucleosome dyads (opaque) and nucleosome footprints (transparent) for the indicated backgrounds at genomic instances of Ume6 recognition motifs. The presence of catalytically active Chd1-Ume6 led to shifted nucleosome dyad peaks (red asterisks) adjacent to predicted Ume6 recognition motifs (blue lines). The Chd1D513N-Ume6 indicates a catalytically inactive Chd1-Ume6 fusion protein containing a Walker B mutation (Hauk et al. 2010). (B, left) Differences in nucleosome dyad signal between Δume6 and Δume6 +[Chd1-Ume6] strains for a 400-bp window centered at permutations of a core GGCGGC motif. Groups 1, 2, and 3 correspond to variations of WNGGCGGCWW, NNGGCGGCNN, and disrupted GGCGGC motifs, respectively. Peak nucleosome movement occurs at the sequence WNGGCGGCWW (asterisk). (Right) Signal differences from GGCGGC-based motifs are compared with other known transcription factor binding motifs from the JASPAR database (Mathelier et al. 2014). Black and red indicate where nucleosome signal increased and decreased, respectively, upon addition of Chd1-Ume6. All ordered motif identities are provided in Supplemental Data File 1. (C) Difference in nucleosome dyad signal between Δume6 and Δume6 +[Chd1-Ume6] strains at all intergenic instances of the WNGGCGGCWW motif, clustered by direction of nucleosome repositioning (left) with average dyad signal within each cluster for individual strains (right).