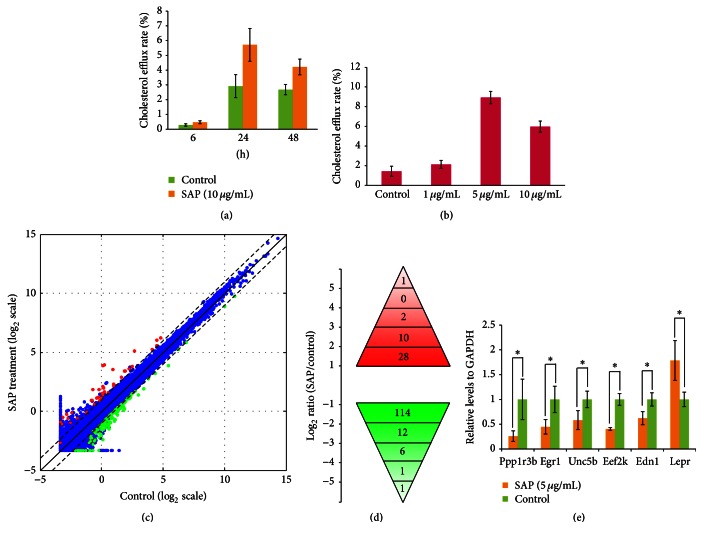
Figure 1.
Systematic identification of genes that are altered in SAP treated cells compared to control cells. (a) SAP enhanced apoA1-mediated cholesterol efflux in a time-dependent manner in macrophages. (b) SAP enhanced apoA1-mediated cholesterol efflux in a dose-dependent manner in macrophages for 24 h. (c) Scatter plot depicting the expression profiles of all genes. Log2 transformed FPKM values from RNA sequencing were used in the scatter plot. We added 1 to FPKM value before log2 transformation to facilitate calculation. Nonchanged genes were shown in blue color while differently expressed genes (fold change > 2 and FDR < 0.05) were denoted in red or green. (d) Distribution of fold change of genes significantly different in SAP treated cells compared to control cells. (e) Validation of RNA sequencing data by quantitative RT-PCR. The GAPDH gene was used as the reference gene for normalization. The statistical significance was tested using unpaired 2-sample t-test. Values were plotted as means ± standard error of the mean (SEM) of triplicate measurements. n = 3. ∗ p < 0.05.