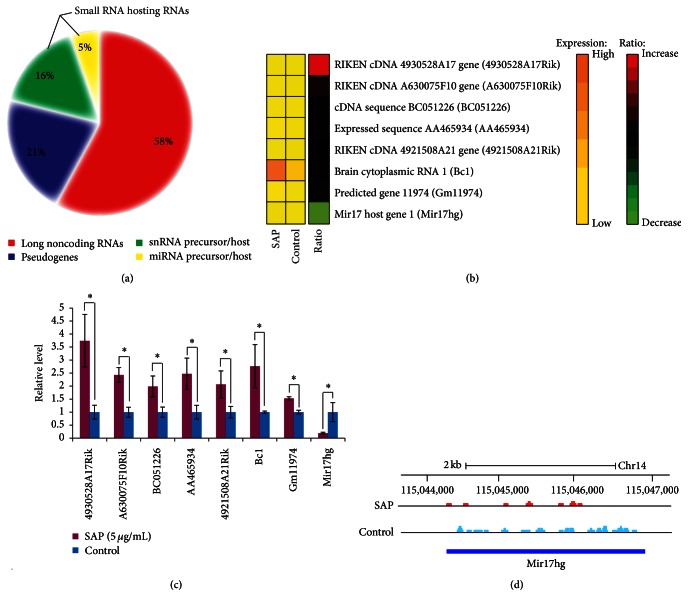
Figure 4.
Changed expression of noncoding RNA genes. (a) Classification of noncoding RNA genes. Pie chart is displayed on the differently expressed 19 noncoding RNA genes. (b) Heat map of the 8 selected long noncoding RNA genes. The first and second columns correspond to the absolute gene expression levels (FPKM values) in SAP group and control group, respectively. The third column of the heat map reports the relative expression levels. Values are color-coded as indicated by the color bars. (c) Quantitative RT-PCR analysis of 8 selected lncRNAs. The GAPDH gene was used as the reference gene for normalization. Statistical significance was tested using unpaired 2-sample t-test. Values were plotted as means ± standard error of the mean (SEM) of triplicate measurements. n = 3. ∗ p < 0.05. (d) RNA sequencing coverage plot of Mir17hg lncRNA. Coverage plot is displayed on the reference genome (UCSC mm9). The upper panel represents expression in SAP group and the lower panel represents expression in control group. For both panels, numbers on y-axis refer to RNA sequencing read-depth at a given nucleotide position.