Figure 1.
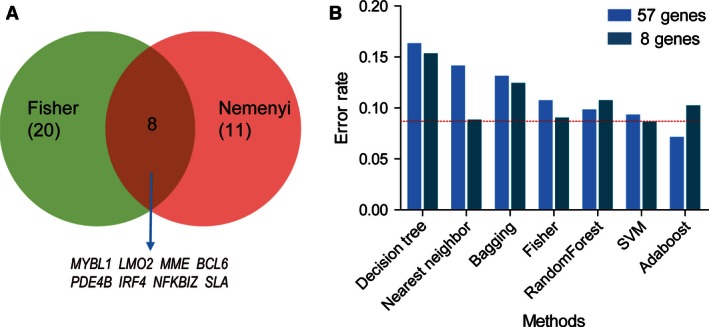
Eight significant genes and support vector machine (SVM) method were selected to construct a new model. (A) Overlaps between genes selected by Fisher discriminant and that by Nemenyi statistical analysis among GCB‐, ABC‐ and unclassified diffuse large B‐cell lymphoma (DLBCL) patients. Venn diagram showed the overlap genes which could stratify the DLBCL patients into GCB and Non‐GCB subtypes. (B) Histogram of the classified error rate of seven machine learning methods between the expression of 57 genes and eight genes with fivefold cross validation in 414 DLBCL patients. The error rate of SVM was the minimum within eight genes model, also smaller than 57 genes model.
