Figure 2.
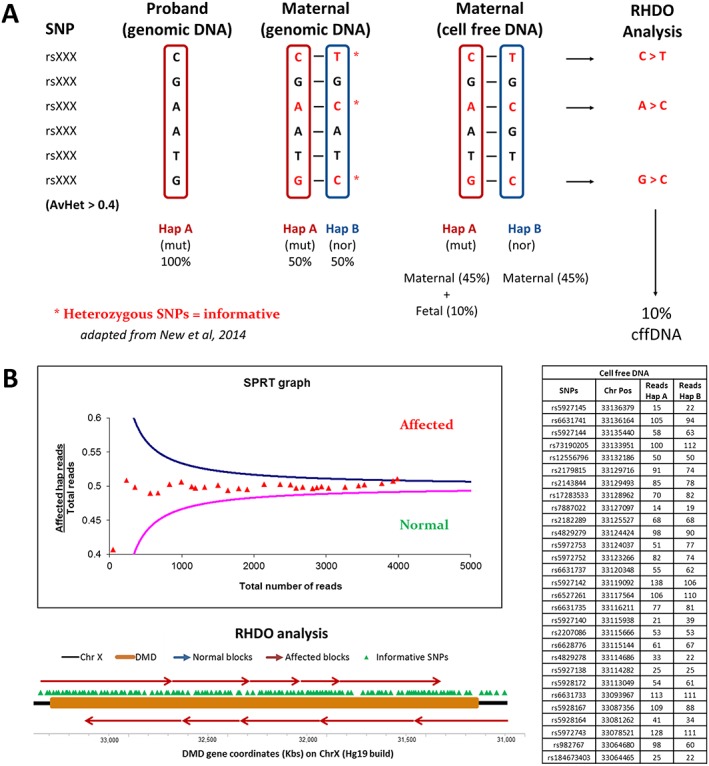
(A) The diagram summarises what is required to conduct relative haplotype dosage (RHDO) analysis for non‐invasive prenatal diagnosis of X‐linked disorders. The cumulative sequencing counts of SNP alleles are used to determine the proband and maternal haplotypes. SNPs that are heterozygous in the mother are informative and are used in the RHDO analysis. The allelic imbalance between the two haplotypes in the maternal cell‐free DNA (cfDNA) is used to calculate the fetal fraction (which has been set at 10% in this case). In this diagram, haplotype A is over‐represented in the maternal cfDNA, indicating that it has been inherited by the fetus. (B) SPRTs are used in RHDO analysis to determine the statistical significance of the allelic imbalance within a haplotype block. Cumulative sequencing counts of SNP alleles from plasma cfDNA (right table) are fed into the SPRT in order of chromosome position until a classification is made. Haplotype blocks are then plotted onto the dystrophin gene and provide the final outcome of the test (diagram). RHDO analysis is conducted from 5′ to 3′ and 3′ to 5′ in order to include all informative SNPs in the analysis and to better estimate the position of recombination sites
