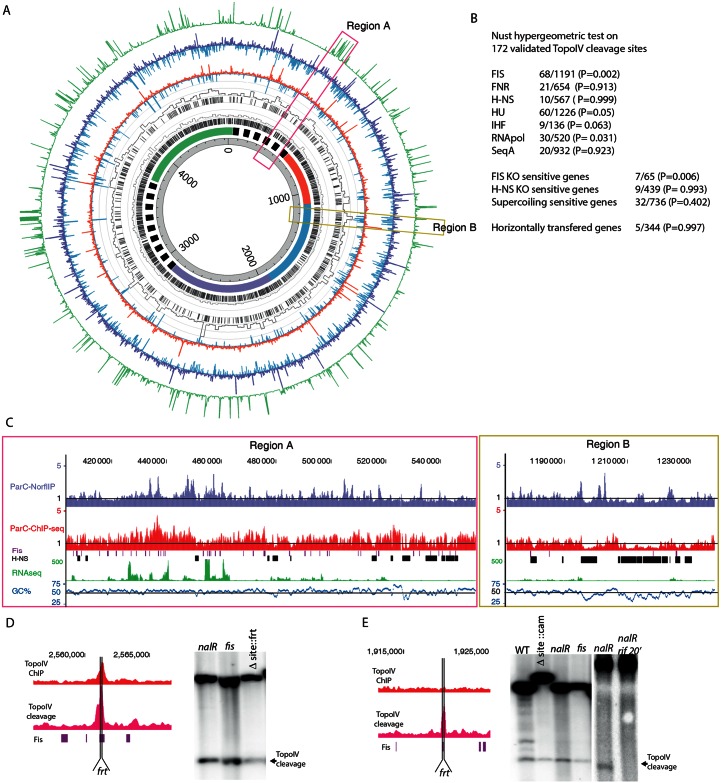
Fig 3. Targeting Topo IV cleavage sites along the E. coli genome.
A) Circos plot of the ParC-Flag Chipseq and ParC-Flag 1 NorflIP experiments. From the center to the outside, circles represent: genomic coordinates, macrodomain map, Fis binding sites in mid exponential phase, % of bases bound by Fis per 20 kb windows of genomic DNA, H-NS binding sites in mid exponential phase, % of bases bound by H-NS per 20 kb windows of genomic DNA [33], ParC-Flag ChIP-seq (depleted regions blue, IP/input <1), ParC-Flag ChIP-seq (enriched regions red, IP/input >1), ParC-Flag 1 NorflIP (depleted regions blue, IP/input <1), ParC-Flag 1 NorflIP (enriched regions red, IP/input >1), gene expression data (RNA-seq results performed in the ChIP-seq and NorflIP conditions). For visualization purpose, the maximum scale of RNAseq data has been fixed to 500 reads which approximately corresponds to the 400 transcription units that were the most expressed (the distribution of read counts scaled from 0 to 30 000). B) Correlation between the localization of Topo IV cleavages and chromatin markers. The NUST [32] hypergeometric test was used to compare Topo IV and chromatin markers localization. The set of 172 validated Topo IV cleavage sites was used. The number of common localizations over the total number of chromatin marker localization is indicated. The P value of a Fisher’s exact test is indicated. C) Genome browser magnifications of the panel A’s pink and yellows regions. Mid log phase Fis and H-NS binding sites are respectively indicated with burgundy and black boxes [33]. D) Magnification of the 2.56 Mb Topo IV binding and cleavage site that overlaps a Fis binding site. The position of the deleted Topo IV site is marked by vertical lines (frt). Southern blot analysis of Topo IV cleavage at the 2.56 Mb locus, in the nalR strain, the nalR strain with a deletion of the Topo IV cleavage and binding site and the deletion of fis. E) Same as D for the 1.92 Mb Topo IV cleavage site. Southern blot analysis of Topo IV cleavage at the 1.92 Mb locus, in the WT, the nalR strain, the nalR strain with a deletion of the Topo IV cleavage and Fis binding site and the nalR strain with fis deletion. The cleavage was also analyzed following a 20 min treatment with rifampicin (rif).