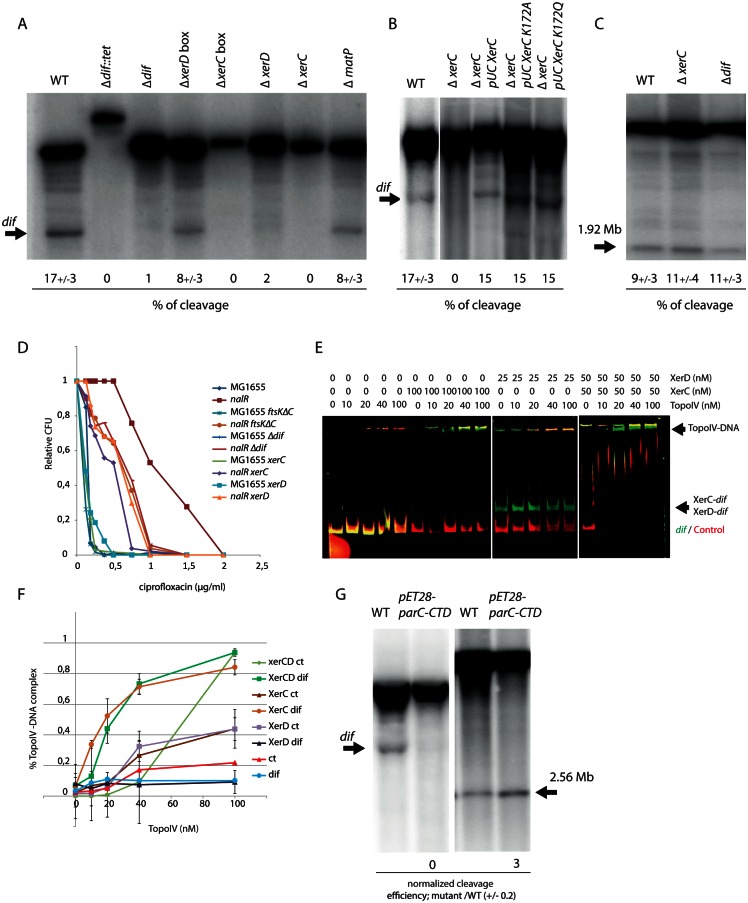
Fig 4. Determinants of Topo IV activity at dif.
A) Southern blot analysis of the Topo IV cleavage at the dif site. Genomic DNA extracted from WT, Δdif::Tc, Δdif, ΔxerD box, ΔxerC box, ΔxerD, and ΔxerC strains was digested by PstI; the size of the fragment generated by Topo IV cleavage at dif is marked by an arrow. The average percentage of cleavage observed in two independent experiments is presented. B) Southern blot analysis of the Topo IV cleavage at the dif site. Genomic DNA extracted from WT, ΔxerC, ΔxerC pUCxerC, ΔxerC pUCxerCK172A, ΔxerC pUCxerCK172Q strains was digested with PstI; the size of the cleaved fragment in dif is marked by an arrow. C) Topo IV cleavage at the 1.9Mb site in the WT, ΔxerC and Δdif. D) Plating of parEts, parEts xerC, parEts xerD and parEts recN mutants at 30 and 37°C. E) Colony Forming Unit (CFU) analysis of the WT and nalR strains deleted for the dif site, the xerC, xerD genes or the C-terminal domain of FtsK in the presence of ciprofloxacin. F) EMSA on a 250 bp CY3 probe containing dif (green) and a 250 bp CY5 control probe (red). The amount of Topo IV, XerC or XerD proteins present in each line is indicated above the gel. G) Quantification of Topo IV EMSA presented in C, data are an average of three experiments. H) Southern blot analysis of Topo IV cleavage at dif and position 1.92Mb in a strain overexpressing the C-terminal domain of ParC.