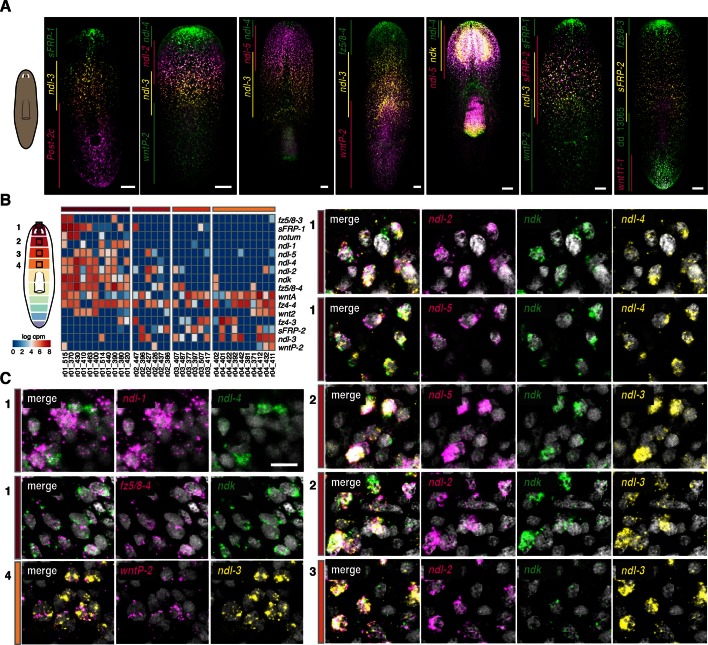
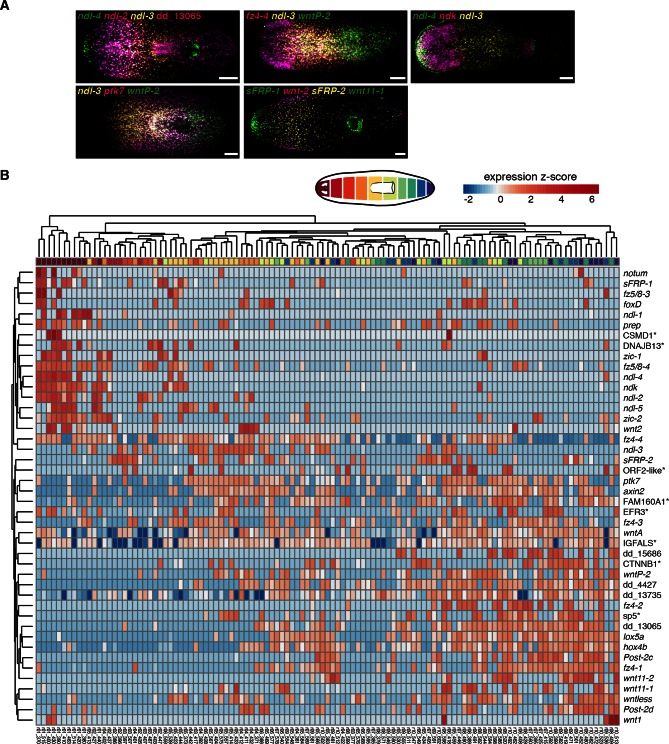
Figure 2. Co-expression of mRGs along the AP axis.
(A) FISH using mRGs maps discrete domains of mRG expression onto the planarian AP axis. Bars on left indicate the approximate extent of the expression domain for each of the genes analyzed. Images are representative of n≥5 animals. Anterior, up. Scale bar, 100 μm. (B) Heatmap shows co-expression of anterior FGFRL and Wnt pathway mRGs in the four anterior regions indicated in the cartoon (1–4). Each column shows expression within a single cell with color bars above indicating the dissected region of origin for the cell. cpm, counts per million (C) FISH using different FGFRL/ndl probes and Wnt pathway mRGs show co-expression in the four regions depicted in the cartoon. Black boxes (1–4) in the cartoon in B show the region imaged for the FISH, as denoted by the number and colored rectangle next to the merged image. Scale bar, 10 μm. Images are representative of n≥5 animals.