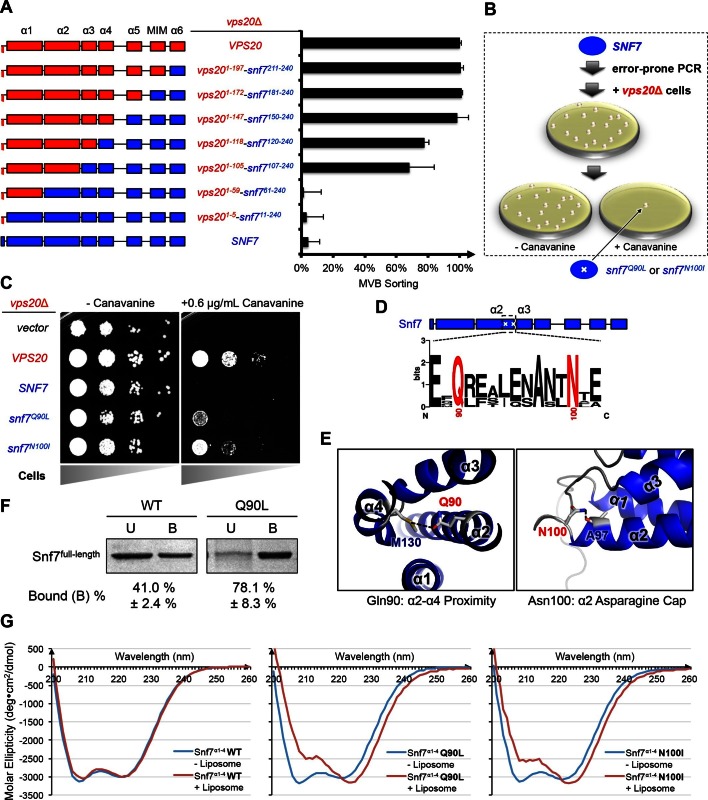
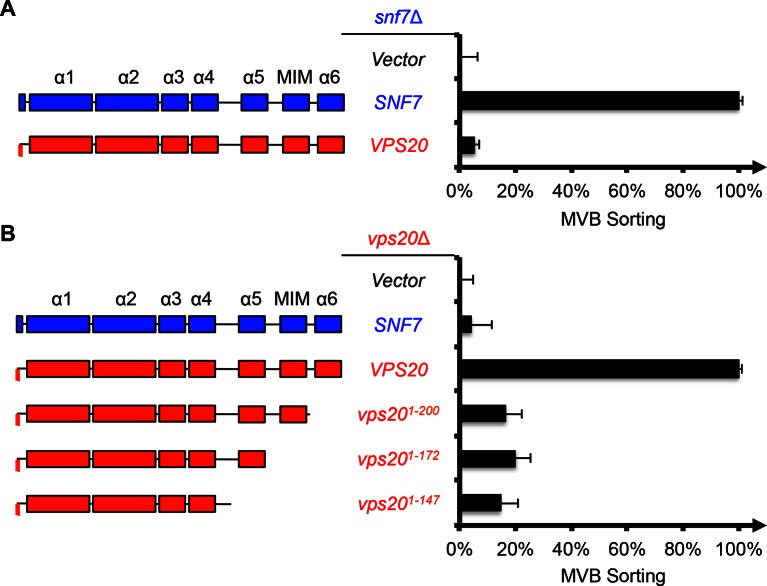
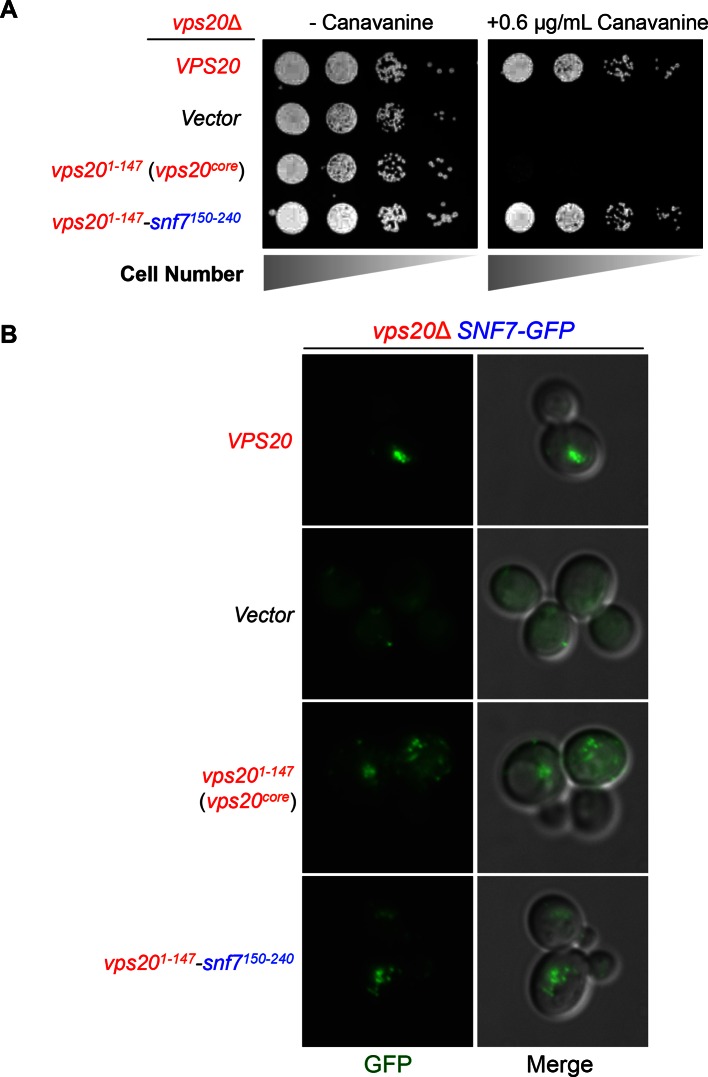
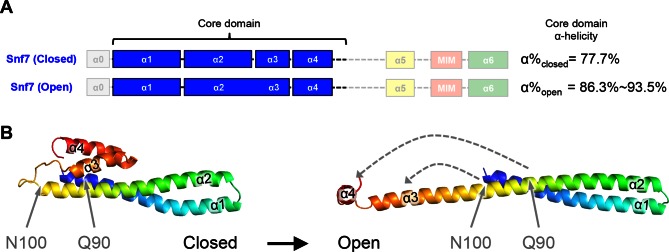
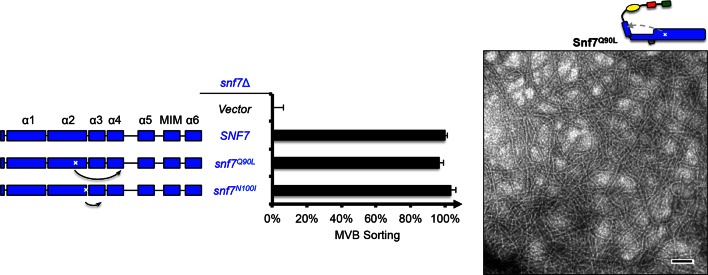
Figure 1. Novel Snf7 point mutations trigger core domain activation.
(A) Domain organization of Vps20(red)-Snf7(blue) chimera (left) and quantitative MVB sorting data (right) for vps20Δ yeast exogenously expressing VPS20, vps201-197-snf7211-240, vps201-172-snf7181-240, vps201-147-snf7150-240, vps201-118-snf7120-240, vps201-105-snf7107-240, vps201-59-snf761-240, vps201-5-snf711-240, and SNF7. Error bars represent standard deviations from 3–5 independent experiments. (B) Screening strategy to identify snf7 suppressors in vps20Δ yeast. (C) Canavanine sensitivity assay for vps20Δ yeast exogenously expressing empty vector, VPS20, SNF7, snf7Q90L, and snf7N100I. (D) Domain organization of Snf7, with the locations of Gln90 and Asn100. WebLogo of protein sequence analysis (Doerks et al., 2002) of Snf7 orthologs from Saccharomyces cerevisiae, Homo sapiens, Mus musculus, Xenopus laevis, Drosophila melanogaster, Caenorhabditis elegans, Schizosaccharomyces pombe. (E) Close-up view of the side chain interactions of Gln90 (left) and Asn100 (right) in a 'closed' Snf7 homology model (Henne et al., 2012). (F) Liposome sedimentation assays of Snf7WT and Snf7Q90L. Liposome-bound (B) proteins and unbound (U) proteins. (G) CD scanning spectra from 200 nm to 260 nm of wild-type Snf7α1–4 (left), Snf7α1–4 Q90L (middle), and Snf7α1–4 N100I proteins with (red) and without (blue) liposomes.