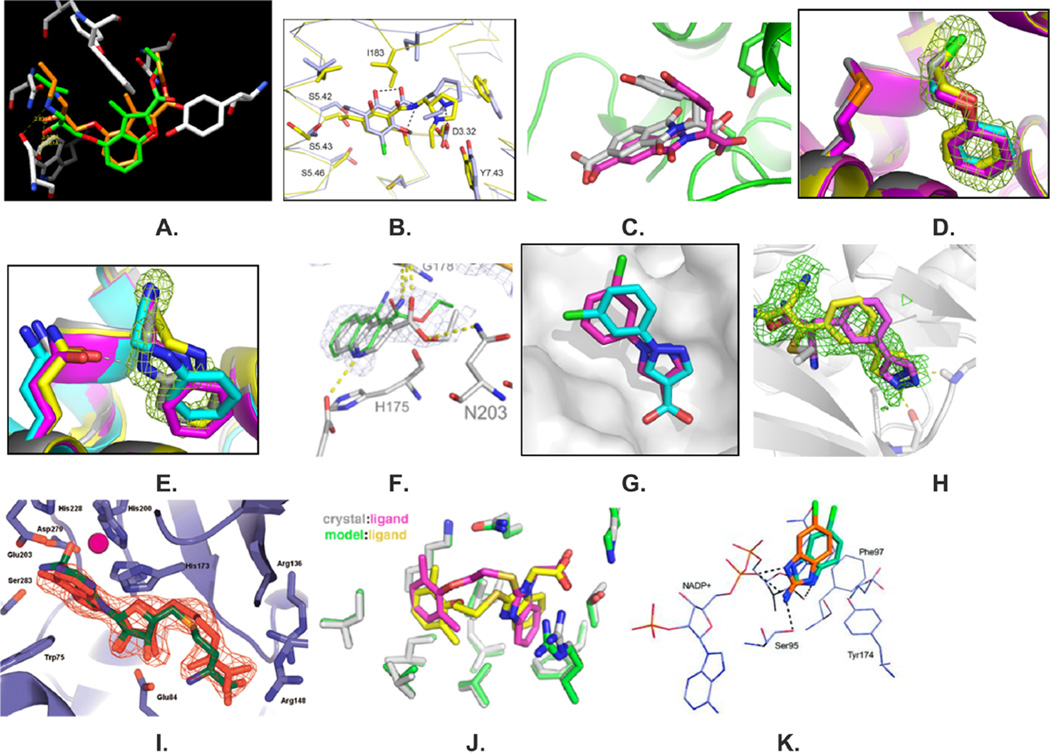
Figure 2.
Superposition of docking-predicted and subsequently determined crystal structures. (A) A 9 nM inverse agonist of the β2 adrenergic receptor discovered by docking (orange) superposed on the crystallographic result (green).49 (B) Docking-predicted structure of eticlopride (light blue) superposed on the crystallographic result (yellow) in the dopamine D3 receptor. Reproduced with permission from Nature Chemical Biology (Carlsson, J.; et al. Ligand discovery from a dopamine D3 receptor homology model and crystal structure; 2011, 7, 769–778);213 Copyright 2011 Macmillan Publishers Ltd. (C) A 30 µM inhibitor of β-lactamase discovered by docking (purple) superposed on its crystallographic structure (white).48 (D, E) Two crystal structures (gray carbons, electron density in wire mesh) superposed on the docking predicted ligands (yellow carbons) and poses refined by postdocking rescoring programs AMBERDOCK (cyan carbons) and PLOP (magenta carbons) for the nonpolar T4 lysozyme L99A and L99A/M102Q model cavities. Reproduced with permission from Journal of Molecular Biology (Graves, A. P.; et al. Rescoring docking hit lists for model cavity sites: predictions and experimental testing; 2008, 377, 914–934);245 Copyright 2008 Elsevier, Ltd. (F) A docked ligand superposed on the crystallographic result from a screen against the model anion cavity in cytochrome c peroxidase W191G/gateless. Reproduced with permission from Nature Chemistry (Fischer, M.; et al. Incorporation of protein flexibility and conformational energy penalties in docking screens to improve ligand discovery; 2014, 6, 575–583);243 Copyright 2014 Macmillan Publishers Ltd. (G) A 40 nM boronic acid inhibitor of AmpC predicted by covalent docking (cyan carbons) superposed on its crystal structure with the enzyme.175 (H) A 370 nM inhibitor of RSK2 kinase predicted by covalent docking (magenta carbons) superposed on its crystal structure with the enzyme (yellow carbons, with Fo – Fc electron density shown in green mesh.175 (I) The docked structure (green carbons) of the high-energy intermediate of S-adenosyl homocysteine superposed on the crystallographic structure of the product, S-inosyl homocysteine (red carbons) in complex with Tm0936. Reproduced with permission from Nature (Hermann, J. C.; et al. Structure-based activity prediction for an enzyme of unknown function; 2007, 448, 775–779);198 Copyright 2007 Macmillan Publishers Ltd. (J) An 8 µM inhibitor of PDK1 showing the docking pose (yellow carbons) superposed on the subsequently determined crystal structure (magenta carbons).234 (K) Modeled binding mode of a 10.6 µM fragment (orange carbons) superimposed with the dominant binding mode determined crystallographically (green carbon atoms).251