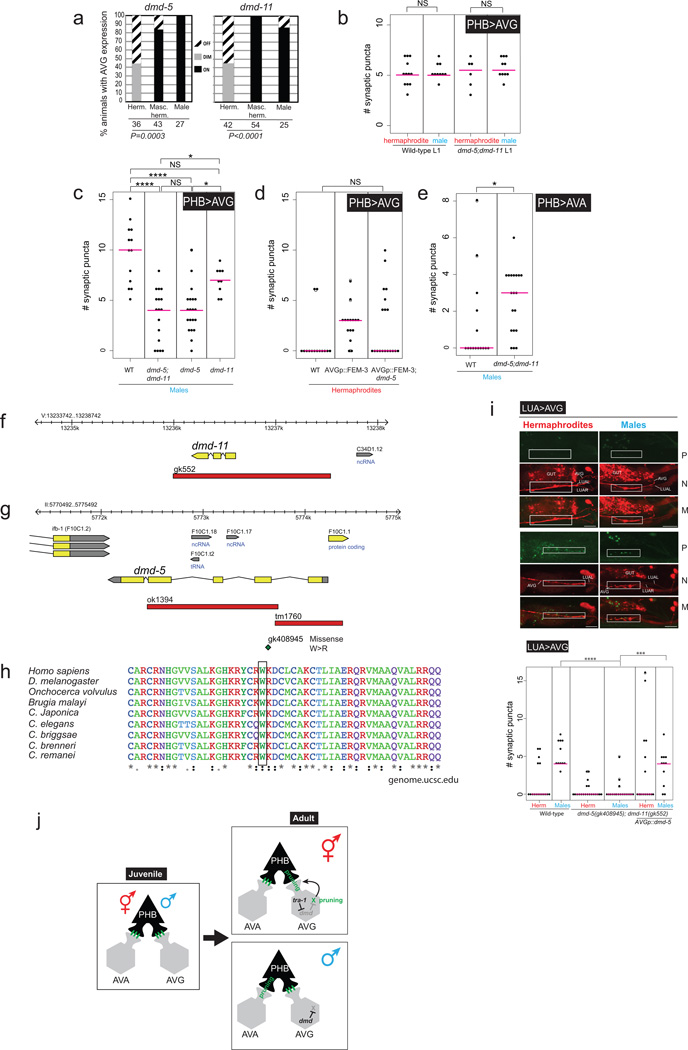
Extended Data Figure 9. dmd-5 and dmd-11 expression, sequence and function.
a: Quantification of dimorphic expression of dmd-5 and dmd-11 in AVG. Expression in hermaphrodites was off or extremely faint. Expression of inx-18p::FEM-3 derepressed dmd-5 and dmd-11 gene expression in hermaphrodite AVGs. Statistics calculated using Fischer’s Exact test.
b: Quantification of the number of PHB-AVG synaptic puncta in L1 dmd-5(gk408945); dmd-11(gk552) double mutants, compared with wild-type L1 animals. At the L1 stage dmd-5 and dmd-11 do not affect PHB-AVG synapses, suggesting they are required for maintenance of mature synapses.
c:dmd-5 single mutants and dmd-5; dmd-11 double mutants display similar alterations in AVG synaptic wiring.
d:dmd-5 mutation suppresses the ectopic PHB>AVG synapses in AVG-masculinized animals.
e: The PHB>AVA connection is non-autonomously partially stabilized in dmd-5; −11 mutants.
f:dmd-11 genomic locus and gk552 deletion location.
g:dmd-5 genomic locus and mutation description. ok1394 location was not curated, to determine location we used the following primers: Forward primer: cagaatgcctgtttctccgtc and Reverse: cactgcttttcccgttcaaac. ok1394 and tm1760 were both found to have an embryonic lethal phenotype that couldn’t be rescued with the genomic locus (data not shown), thus we searched for single point mutations of the “million mutation project”43gk408945 is a missense substitution mutation of W54 to R, located in the second exon. Genomic analysis revealed that this mutation lies within the conserved DM domain (e.), with perfect conservation across evolution. The DM domain is an intertwined zinc-containing DNA binding module. The DM domain binds DNA as a dimer, allowing the recognition of pseudopalindromic sequences 44.
h: DM-domain sequence conservation and location of gk408945 mutation. Conservation and multiple sequence alignment were done using UCSC Genome Browser (genome.ucsc.edu) and ClustalW.
i:dmd-5 and dmd-11 are required for maintenance of AVG synapses. Fluorescent micrographs and quantification of synaptic puncta of LUA>AVG. Region of neurite overlap and observed synaptic puncta marked with white boxes. The P, M and N letters next to fluorescent images denote labeling of Puncta, Neurite and Merge, respectively.
In b, d , e and i statistics were calculated using the nonparametric Mann-Whitney test. In panel c statistics were calculated using Kruskal-Wallis test with Dunn’s multiple comparison test. ****P < 0.0001, ***P < 0.001, *P < 0.05, NS; Not significant. Magenta horizontal bars represent the median. When using a parametric t-test, there is also a significant difference for the LUA>AVG synapse between dmd-5;dmd-11 mutant hermaphrodites and dmd-5;dmd-11 mutant hermaphrodites that over express DMD-5 (*P < 0.05).
j: Summary of data. TRA-1 and DMD proteins are commonly thought to work as transcriptional repressors 22. Since dmd-5/11 are already dimorphically expressed in AVG in embryos and L1 stage animals (not shown in this schematic), there must be other timer mechanisms that control the onset of pruning. For example, DMD-5/11 may work together with a regulatory factor of the stage-specifically acting heterochronic pathway. Furthermore, we hypothesize that other neurons, such as the AVA neuron, may have its own complement of sex-specific dmd genes that control pruning.