Figure 3.
Chemotaxis-Based Radial Intercalation Hypothesis and Computer Simulation
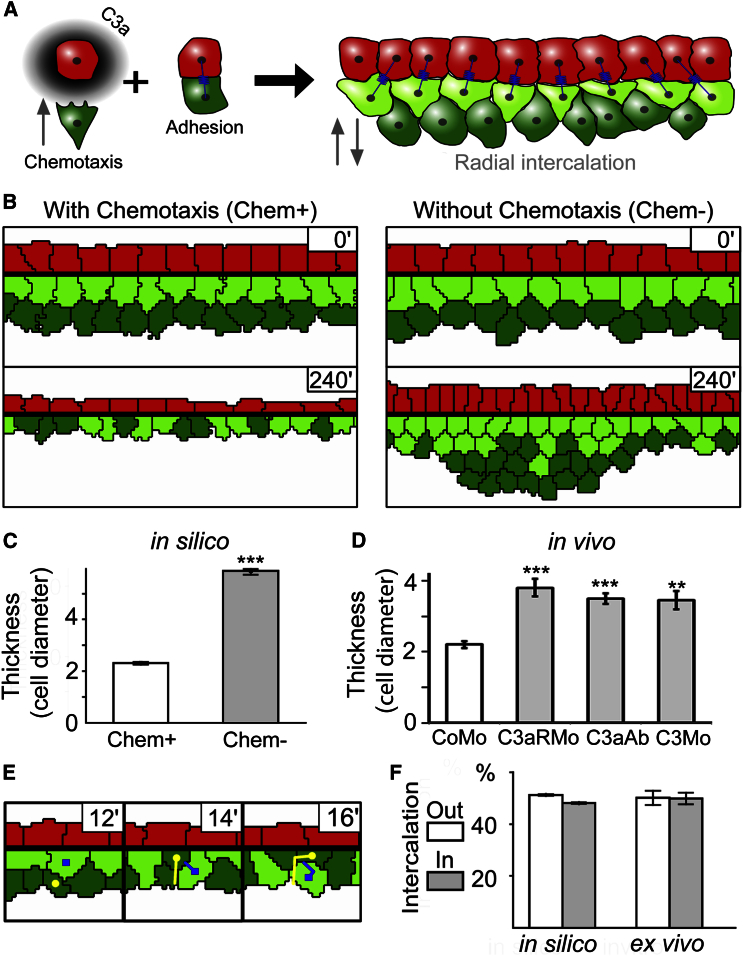
(A) Hypothesis: chemotaxis of DCs (green) toward SCs (red) via C3a drives RI and consequent tissue thinning.
(B) Computer simulations of the hypothesis showing the initial (0′) and final (240′) cell configurations with chemotaxis (Chem+) and without chemotaxis (Chem-). Coloring as in Figures 1A and 1B, for visual aid only.
(C) Tissue thickness, measured in units of cell diameter after epiboly (20 μm), is significantly increased in simulations without chemotaxis (n = 30).
(D) In vivo tissue thickness, measured in units of cell diameter after epiboly (20 μm), is significantly higher in embryos with either impaired C3 receptors (C3aRMo, n = 62) or C3 ligands (C3aAb, n = 35; C3Mo, n = 62) compared with control embryos (CoMo).
(E) Prediction of bidirectional intercalation. In silico, DCs intercalate both toward the secreting SL, driven by chemotaxis, and against the chemotactic gradient away from the SL, driven by volume exclusion.
(F) In silico DC movement toward the SL (Out) is only slightly more frequent than movement away from it (In). Direction of DC intercalation measured in the intercalation assay (ex vivo, see Figure 2) confirms the model's prediction.
Error bars: SD, significance ∗∗p < 0.01, ∗∗∗p < 0.001. See also Figure S6.