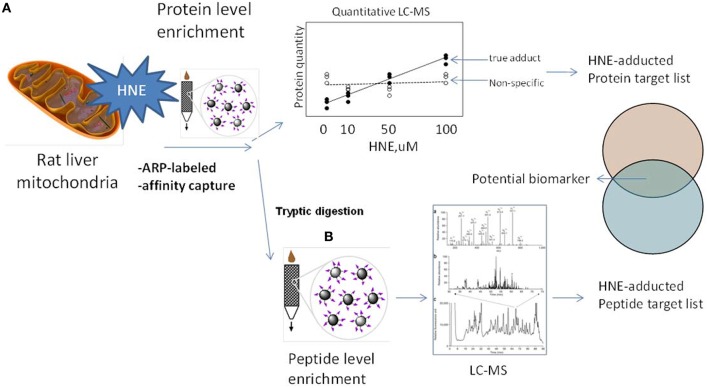
Figure 2.
Experimental design and workflows used in this study. To explore the protein targets of HNE mitochondrial samples were exposed to HNE, captured, identified, and quantified by implementing two complementary capture strategies: (A) at the protein level and (B) at the peptide level. We used the ARP probe to label HNE-protein adducts. In strategy (A) we performed affinity capture at the protein level. After trypsin digestion, samples were analyzed by label-free quantification approach to identify and quantify protein targets that increase with HNE concentration to obtain a preliminary target list. In strategy (B) we applied the affinity capture step at the peptide level, i.e., tryptic digestion of samples were performed prior to the enrichment of HNE-adducted peptides. By this approach, another list of targets with site-specific information was obtained. Those found in both lists were proteins that showed a concentration-dependent response as well as known modification sites. These protein targets of HNE can serve as biomarker candidates useful for future monitoring and/or diagnostic purposes in preclinical models of inflammatory hepatic diseases.