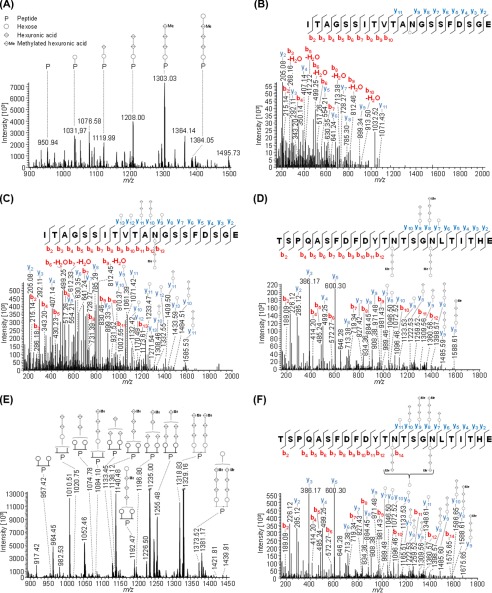
FIGURE 5.
Mass spectrometric identification of PilA2 N-glycosylation. Peptides of GluC-digested proteins from CsCl density gradients obtained from WT cells were analyzed by MS. IS-CID was applied for A and B (80 V) and E (60 V); therefore, the N-glycan composition could be analyzed by a series of neutral losses from the N-glycosylated peptide ion (P) in the MS1 spectrum (A and E) and is depicted with symbols: hexose (○), hexuronic acid (♢), methylated hexuronic acid (♢Me). A pentasaccharide glycan could be identified (A), and the corresponding N-glycopeptide sequence was analyzed by further HCD fragmentation of m/z 1031.97 (charge 2) corresponding to the peptide + hexose leading to the identification of ITAGSSITVTAN@GSSFDSGE (B) from PilA2. The pentasaccharide N-glycosylation could be confirmed by HCD fragmentation of intact glycopeptides without IS-CID for PilA2 peptides ITAGSSITVTAN@GSSFDSGE (C, precursor m/z 1384.55, charge 2) and TSPQASFDFDYTN@TSGN@LTITHE (D, precursor m/z 1427.86, charge 3), which contains two glycosites. The two glycosites from TSPQASFDFDYTN@TSGN@LTITHE can harbor N-glycans of different length as observed in the MS1 spectrum (E) after applying IS-CID. The presence of a tetrasaccharide, lacking the terminal hexose, and a pentasaccharide on the same peptide was confirmed without IS-CID by HCD fragmentation of the triply charged precursor m/z 1075.44 (F). It could not be specified which of the two glycosites was modified by the tetra- and pentasaccharide, respectively. The observed b-ions (red) and y-ions (blue) are indicated in the spectra as well as the fragmentation schemes, including attached glycan fragments. Some peaks (slanted parallel bars) are not shown with their full intensity for better visualization (n = 2).