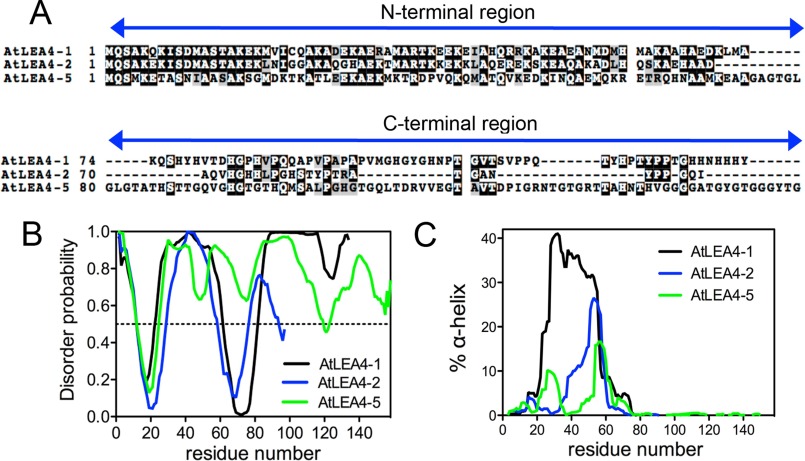
FIGURE 1.
In silico structure prediction of Arabidopsis group 4 LEA proteins. A, sequence alignment of Arabidopsis group 4 LEA proteins showing N-terminal conserved and C-terminal variable regions. B, AtLEA4-1 (black), AtLEA4-2 (blue), and AtLEA4-5 (green) structural disorder levels using PONDR predictor. According to this algorithm, proteins with disorder probability values above 0.5 are considered highly disordered. C, percentage of α-helix predicted using AGADIR for AtLEA4-1 (black), AtLEA4-2 (blue), and AtLEA4-5 (green).