Fig. 7.
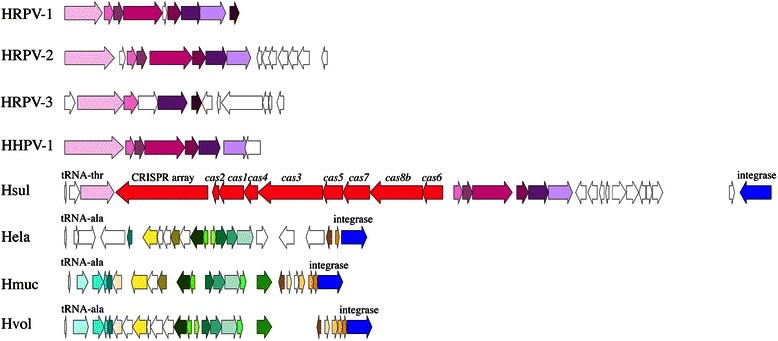
Comparative genome map with genome alignment results. Relative lytic pleomorphic viruses HRPV1, HRPV2, HRPV3, HHPV1 have seven core genes (colored in shades of magenta) that are shared between them and a prophage 1 from ‘Halanaeroarchaeum sulfurireducens’ M27-SA2. Relative prophages of Haloferax Hela, Hmuc, Hvol (coloured in shades of green) have tRNA and XerC/D integrase/recombinase flanking their genomes. The same flanks has the genome of ‘Halanaeroarchaeum sulfurireducens’ M27-SA2 prophage Hsul. CRISPR array and cas genes are colored in red. XerC/D integrase/recombinase is colored in blue. The same colors of the genes in genomes represent homologous genes. Short designations used: HRPV1, Halorubrum pleomorphic virus 1; HRPV2, Halorubrum pleomorphic virus 2; HRPV3, Halorubrum pleomorphic virus 3; HHPV1, Haloarcula pleomorphic virus 1; Hsul, ‘Halanaeroarchaeum sulfurireducens’ M27-SA2 prophage 1; Hela; Haloferax elongans contig AOLK01000011 [38]; Hmuc, Haloferax mucosum contig AOLN01000011 [42]; Hvol, Haloferax volcanii chrom1 [42]
