Fig. 4.
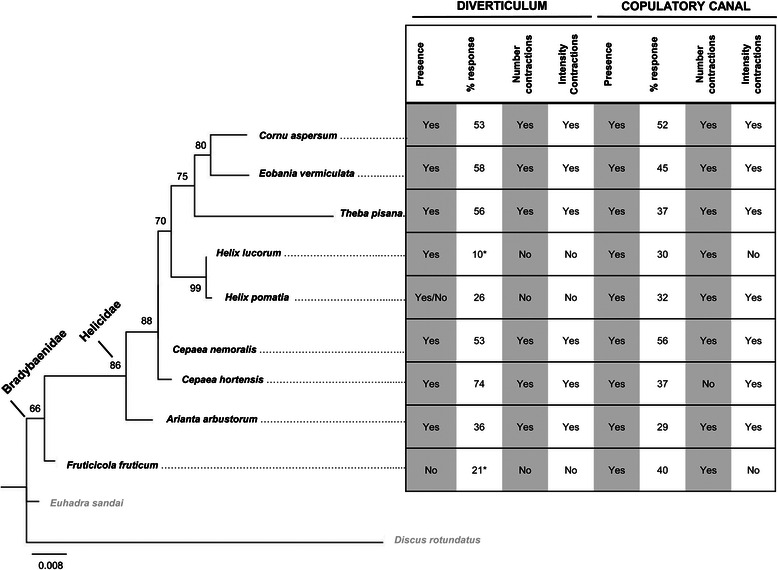
Summary of the results and phylogeny of the species used in this study. The ML phylogenetic tree is shown on the left side. The phylogeny is based on 726 nucleotide sites using Tamura-3-parameter model with a gamma distribution. Bootstrap values above 50 % support are given to the nodes (1000 replications). The length of branches refers to the estimated number of changes occurred between nodes (see scale bar). Discus rotundatus is the outgroup and Euhadra sandai was chosen to support the Bradybaenidae. On the right side, a summary table of the results for both diverticulum and copulatory canal is shown (each row corresponds to each species used in this study). The first column of each organ shows whether it is present (Yes), absent (No) or facultative (Yes/No). The second column shows the percentages of response to the mucous gland extracts for each species. For the focal species Cornu aspersum, the average response of the two time points the mucus was tested is given (once at the beginning and once at the end of each experimental day). The asterisks refer to the significant Chi-square test when that species was compared to the focal species. The last two columns refer to the results shown in Fig. 3 indicating whether the response was similar to the one of the focal species (Yes) or not (No)
