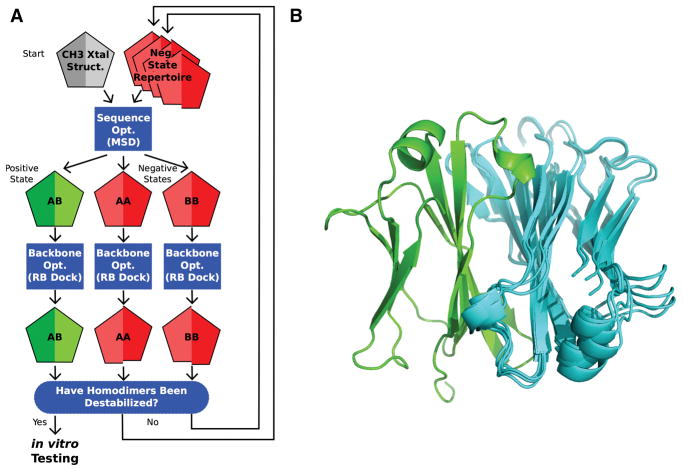
Figure 1. Negative State Repertoires (NSRs).
A) The iterative design process begins by running the multistate design (MSD) sequence optimization executable, feeding in the wild type CH3 homodimer backbone from the 1L6X crystal structure (grey pentagon; each chain of the homodimer is represented by half a pentagon). At its completion, MSD outputs models of the AB heterodimer positive state (green pentagon), and the AA and BB homodimer negative states (red pentagons), threaded onto the backbone of the input structures that produce the lowest-energy. MSD, in trying to destabilize the homodimers, typically introduces collisions across their interfaces; however, these collisions often disappear if the interfaces are allowed to relax. Such false collisions dupe MSD into outputting sub-optimal sequences. Next, backbone optimization through rigid-body docking (RB dock) identifies nearby, low-energy conformations. If the homodimer-backbone conformations it finds (shifted red pentagons) show favorable binding energies, then these structures are added to a NSR for use in subsequent iterations. B) The seven CH3 conformations in the NSR of design 20.8, aligned on chain A (green). Alternate rigid-body orientations of chain B (cyan) show the conformational variability within the space of structures that gave favorable binding energies.