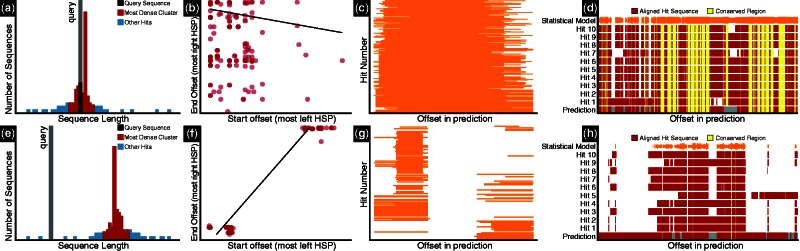
Fig. 1.
Contrasting GV graphs: (a), (e) sequence lengths; (b), (f) HSP offsets; (c), (g) overviews of hit regions; (d), (h) conserved regions. Graphs (a–d) were produced with a sequence for which GV detected no problems. The other graphs show typical problems: (e) query is short; (f), (g) query sequence is a fusion of unrelated genes; (h): query includes sequence absent from first 10 hits