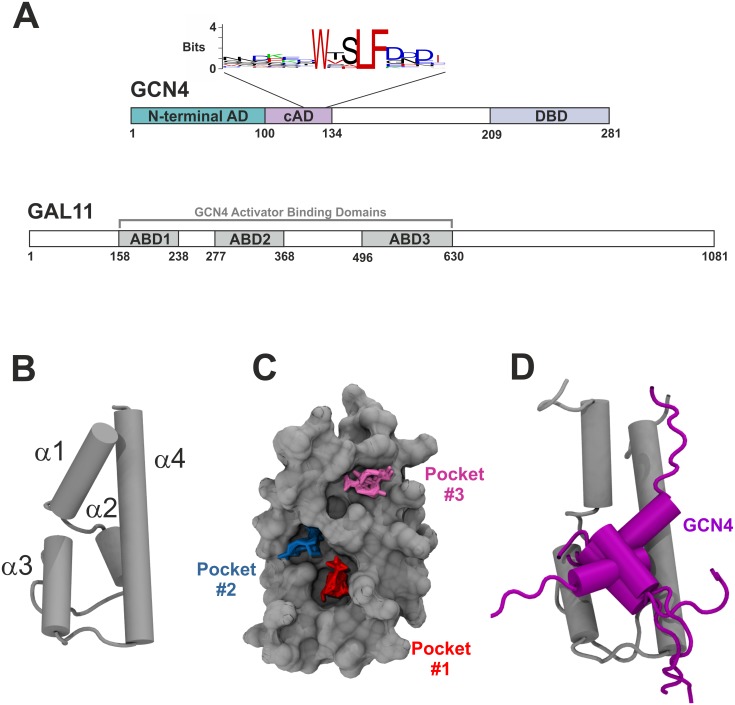
Fig 2. Transcriptional activation of GCN4 via Mediator GAL11.
A. Domain organization of GCN4 (top) and GAL11 (bottom). The positions of the two tandem activation domains (N-terminal AD, central AD [cAD]) and the DNA-binding domain (DBD) of GCN4 are illustrated. A sequence logo based on GCN4 orthologs from 29 different species (see S1 Text for sequence alignments) shows the absolute conservation of three large hydrophobic residues (corresponding to W120, L123 and F124 in Saccharomyces cerevisiae GCN4). GAL11 contains three domains,ABD1, ABD2 and ABD3, that each can bind GCN4 independently. B. Structure of GAL11-ABD1. Cartoon presentation of the uncomplexed structure showing positions of the four separate α-helices (α1 to α4 from N- to C-terminus). C. Surface view showing the location of three deep pockets that display a capacity for binding hydrophobic side chains of partner proteins. Liquorice representations of probe clusters of small organic compounds are shown fitted into three distinct pockets (red = "Pocket#1"; blue = "Pocket#2"; magenta = "Pocket#3") mapped by a computational solvent mapping program [32]. The model shows GAL11-ABD1 residues 164–233. D. Model of the GCN4-cAD bound to GAL11-ABD1. The GAL11-ABD1 and GCN4 structures are shown in silver and purple, respectively. Five different positions, reflecting orientations at 200 ns intervals of aMD_no1, are drawn for the GCN4 cAD in cartoon representation.