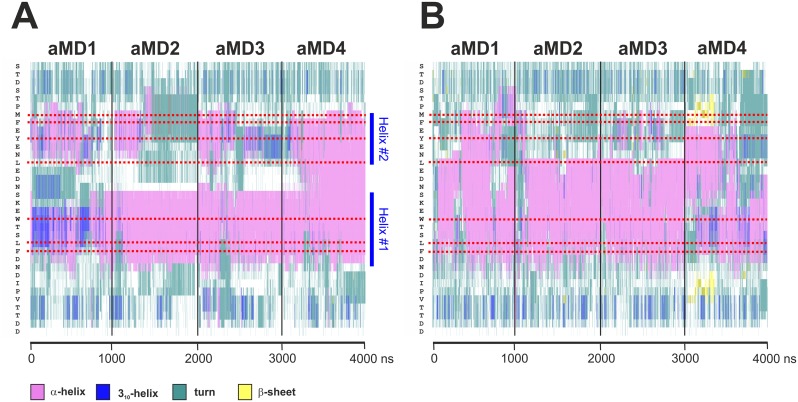
Fig 8. α-helicity of coactivator-bound and free GCN4-cAD.
The trajectories of four aMD simulations (GCN4_aMD_no1 to no4) were combined to allow comparisons across the entire range. The secondary structure is color-coded (pink: α-helix; dark blue: 310 helix; turquoise: turn; white: coil; yellow: β-sheet). The amino acid sequence is on the vertical scale (N-terminus at top; the position of key residues is marked by red dotted lines) and time in nanoseconds of aMD on the horizontal scale. A. Secondary structure analysis of the GCN4-cAD bound to GAL11-ABD1. Except for aMD_no1 (GCN4_aMD_no1), which shows the presence of 310 helices at the beginning of the simulation, all other aMDs are almost exclusively α-helical in the region including the bulky hydrophobic residues W120, L123 and F124. There is evidence for relatively stable N- and C-terminal borders (indicated by dotted dark blue lines at residues S117 and D125, respectively), especially in aMD_no2 and 3 for the central helix ("Helix #1") that encompass W120, L123 and F124. In addition, there is evidence for the presence of an additional transient α-helix ("Helix #2") surrounding residues M107, F108, Y110 and L113. B. Secondary structure analysis of de novo folded GCN4-cAD. There is a widespread formation of short-lived α-helices at different positions and lengths that include residues W120, L123 and F124.