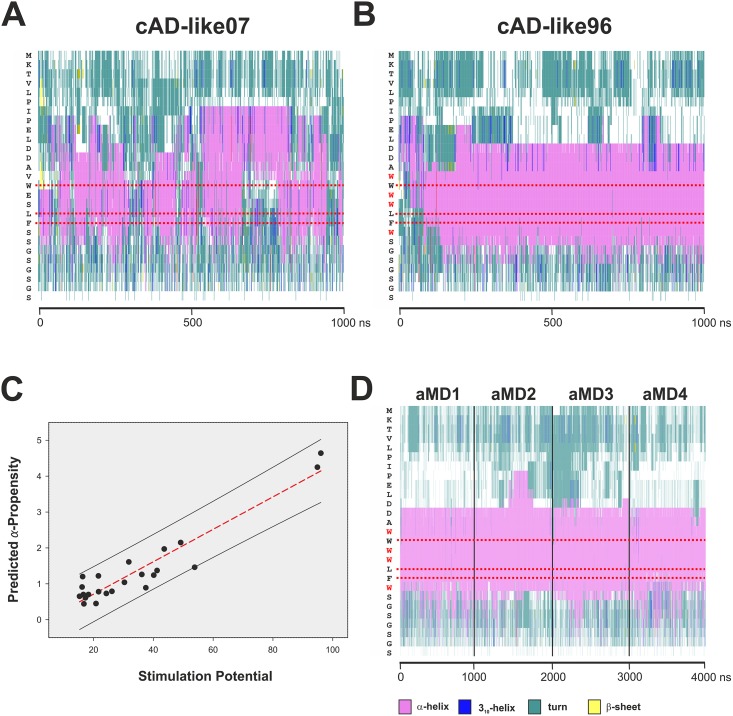
Fig 9. Secondary structures of cAD-like07 and cAD-like96.
The complete sequence of the simulated polypeptide chain is displayed vertically on the left of each graph (Panels A, B and D). The horizontal axis represents time of aMD simulation in nanoseconds. Horizontal red dotted bars mark the positions of the conserved hydrophobic residues. A color code for secondary structure is shown beneath Panel D. A. Formation of α-helical structures during aMD simulation of cADlike07, displaying poor transactivation potential. B. Formation of α-helical structures during aMD simulation of cADlike96, with the highest observed transactivation potential. Note the more extensive formation of stable α-helices and their long-term stability in cADlike96 in comparison to cADlike07. Also, while the N-terminal boundary of the helical structures in cADlike96 is relatively sharply defined (encompassing two aspartic acid residues), the boundary in cADlike07 is much less stable and extends further N-terminal (proline residues). A comparison of the two graphs shows that there are major differences between the two cADlike domains in the extent of α-helical content and stability, as well as the position and stability of the boundaries of these helices. C. Post hoc correlation of α-helicity with transactivation potential. The α-helical propensity (vertical axis) of the cADlike sequences described in Warfield et al. (Table S2 in [42]) was predicted with the Agadir algorithm (http://agadir.crg.es [44]) and plotted against their corresponding transactivation potential (-fold induction of ARG3 mRNA; black dots). The red-dotted line marks the first-order linear regression (r² = 0.89) and the black lines demarcate the 95% prediction interval (including all data points). D. Secondary structure analysis of cAD-like96 bound to GAL11-ABD-1. The trajectories from four independent one microsecond aMD simulations (GAL11-ABD1/cAD-like96_aMD_no1 to no4; Table 1) were combined. The cAD-like96 displays stable α-helicity and helical boundaries. The conserved bulky hydrophobic residues (W94, L97, and F98; highlighted with red horizontal dotted lines) are more than 99% embedded with an α-helical context.