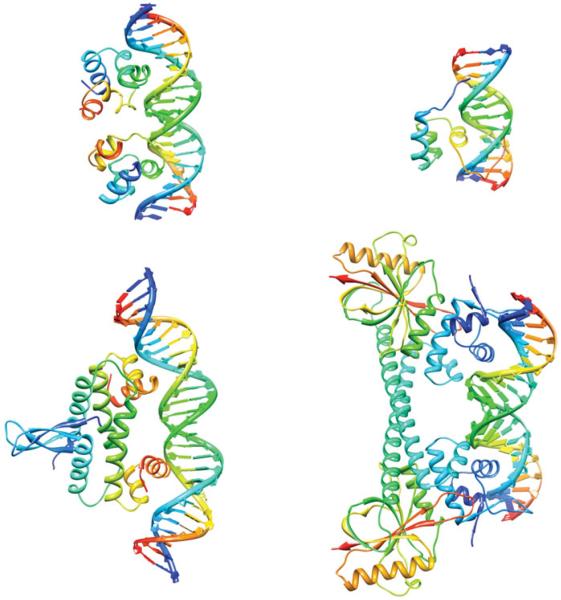
Fig. 1.
Supercoiling and protein–DNA interactions. Top left: the bacteriophage 434 repressor (PDB ID: 3CRO.pdb) has an enhanced affinity for overwound DNA (Koudelka, 1998). Top right: Hin recombinase (PDB ID: 1HCR.pdb) will only bind to a site containing a CAG/CTG triplet when the DNA is supercoiled (Bae et al. 2006). Bottom left: the FIS protein (PDB ID: 3JRE.pdb) is both a transcription factor and a nucleoid-associated protein that constrains negative supercoils (Stella et al. 2010). Bottom right: MerR (PDB ID: 1R8E.pdb) is a bacterial repressor that on binding Hg (II) activates mercury resistance genes by untwisting the DNA-binding site (Ansari et al. 1992).