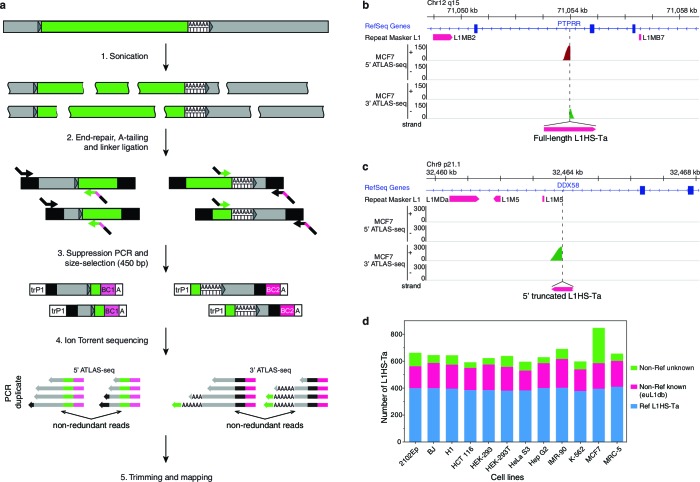
Figure 2. The genetic landscape of L1HS-Ta insertional polymorphisms in 12 human somatic cell lines.
(a) Principle of the ATLAS-seq procedure. The subsequent in silico steps are described in Figure 2—figure supplement 1a. (b–c), Modified IGV genome browser views (Thorvaldsdóttir et al., 2013) of two non-reference polymorphic L1 instances detected in MCF7 cells (b, full length L1, note the two adjacent 5'- and 3'-ATLAS-seq peaks; c, truncated L1). (d) L1HS-Ta insertions found in the various cells of the studied panel. See also Figure 2—figure supplement 1.