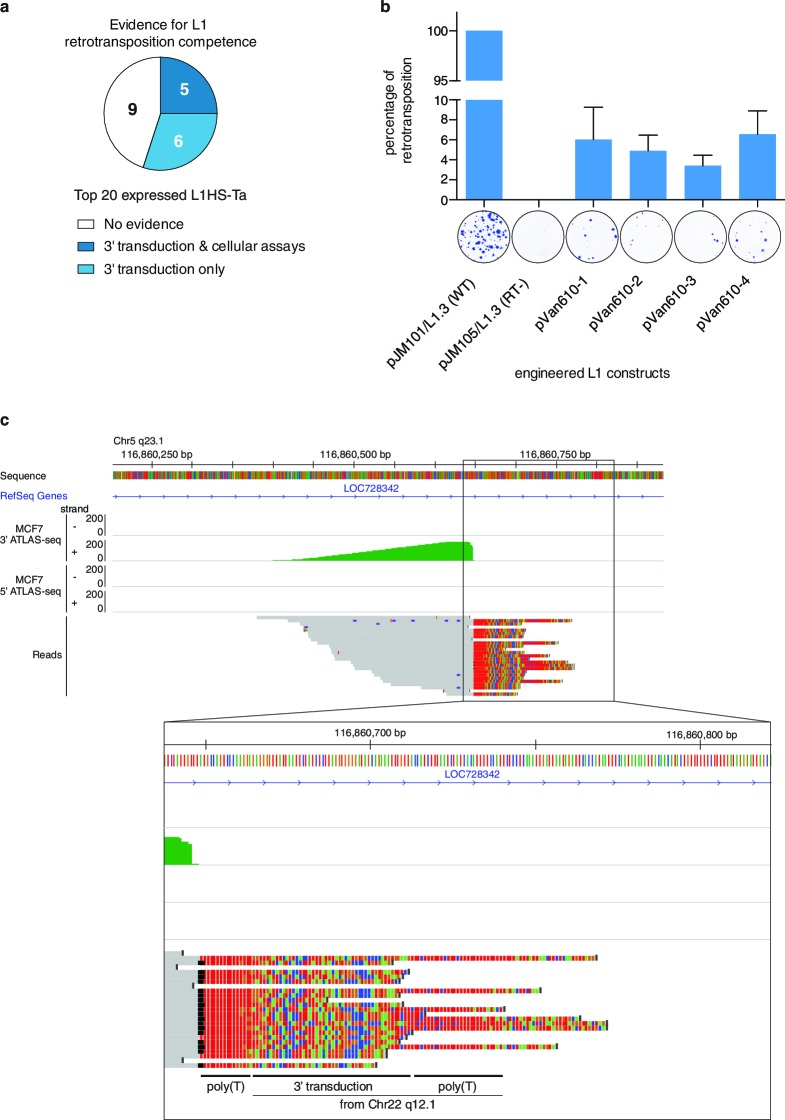
Figure 5. Evidence of retrotransposition capability for selected L1HS-Ta copies.
(a) Evidence of retrotransposition competence for the top 20 most expressed L1 copies across all cell lines analyzed. Cellular assays refer to retrotransposition cellular assays of plasmid-borne L1 instances, whose expression is driven by either the native L1 5’ UTR alone (Brouha et al., 2003) or supplemented by a strong CMV promoter ([Beck et al., 2010] and Figure 5b). These assays measure L1 intrinsic biochemical activity, independently of their actual expression in their genomic context. Three-prime transduction refers to the existence of progeny copies containing a 3' transduction, which can be traced back to the original locus and reflect a retrotransposition event. (b) Retrotransposition assay in cultured cells for MCF7 L1 copy EXP_ID_0447 (NEDD4 locus). A full length transcribed L1HS-Ta copy present in the genome of MCF7 cells was subcloned by PCR in an expression vector containing a reporter gene to measure retrotransposition activity and generated four independent clones (pVan610-1 to -4). In transfected HeLa cells, de novo retrotransposition events of engineered L1 copies lead to the introduction of a functional genomic copy of the neomycin phosphotransferase gene, which expression confers resistance to G418. Resistant foci were stained and counted to monitor retrotransposition activity compared to the positive (pJM101/L1.3, wild type L1HS-Ta) and negative (pJM105/L1.3, mutant L1HS-Ta) control conditions. The value of G418 resistant colonies obtained with the positive control was set to 100%. A picture of a representative well with stained colonies is displayed for illustrative purposes under each bar of the graph. The average value of three biological replicates is displayed with error bars corresponding to the standard deviation among the three biological replicates. (c) Detection of 3' transductions in ATLAS-seq data. This in silico screen identifies L1HS-Ta copy (progeny element) with ATLAS-seq clusters containing reads with non-aligning subsequences (soft-clipped), which uniquely map downstream and adjacent to another full length L1HS locus (progenitor element). The panel shows a genome browser view of such a 3' transduction, originating from a full length L1HS-Ta in the TTC28 gene (22q12.1). The soft-clipped region of the reads is shown in color (base code: T, red; A, green; C, blue; G; orange). As expected, the transduced region is flanked by 2 poly(A) tails (poly(T) here since it is located on the reverse genomic strand).