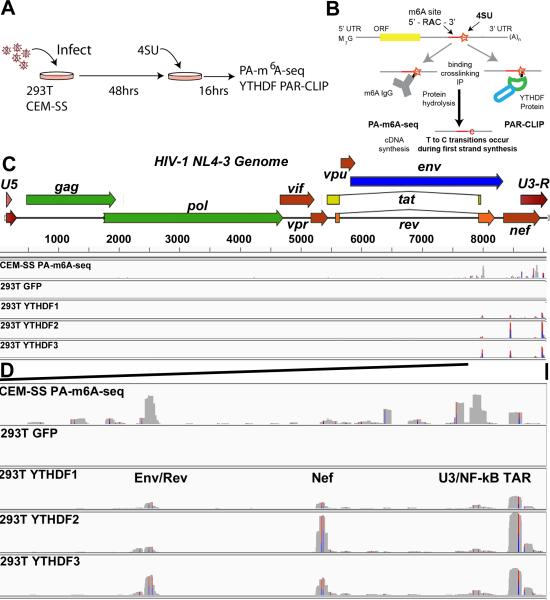
Figure 1. m6A site discovery in HIV-1 isolate NL4-3.
(A) Overview of the general PAR-CLIP experimental design. (B) A schematic of the PA-m6A-seq and PAR-CLIP site discovery strategy is depicted. A typical transcript containing an m6A editing site is shown with an incorporated adjacent 4SU molecule (orange star). Upon binding, the m6A specific antibody or a host YTHDF reader protein is crosslinked to the 4SU. T>C transitions are generated from crosslinked 4SU during reverse transcription/cDNA synthesis. (C) PA-m6A-seq and PAR-CLIP were performed 64 h after infection with VSV-G pseudotyped HIV-1 strain NL4-3. Shown are the entire genome coverage tracks for PA-m6A-seq in CEM-SS cells, and then the FLAG-GFP control and YTHDF1, 2 and 3 tracks in 293T cells. (D) Expanded view of the 3'UTR region of HIV-1 containing the detected m6A editing sites. This ~1.4 kb region extends from the second coding exon of Rev to the end of the R region. Red/Blue bars indicate sites of T>C conversions. Reads are aligned to an HIV-1 genome that begins with the U5 region and ends with U3-R to avoid repeat alignments. The PA-m6A-seq has a Y axis of 0-200 reads, and all others are depicted with Y axes of 0-900 reads. For related data, see Fig. S1.