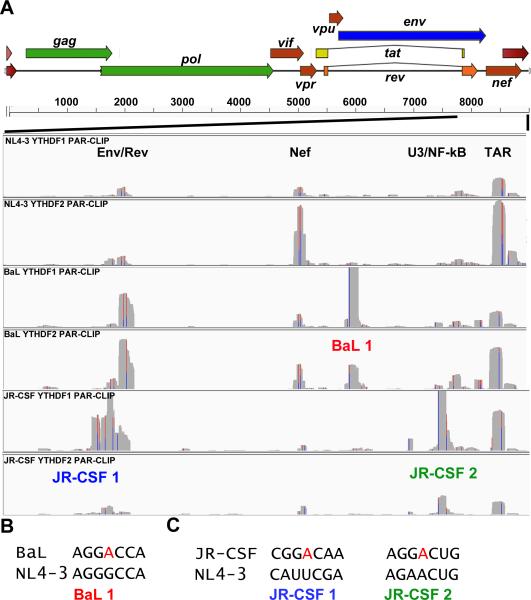
Figure 2. m6A site discovery using primary HIV-1 isolates BaL and JR-CSF.
(A) YTHDF1 or YTHDF2 PAR-CLIP binding clusters were mapped for HIV-1 isolates NL4-3, BaL and JR-CSF for the 3’ region of the HIV-1 genome from the second exon of Rev to the end of the R region, as indicated. The three novel YTHDF protein binding clusters discovered for these two viruses are annotated below the relevant track. The Y axes for these alignments are NL4-3: 0-900 reads, BaL: 0-2000 reads, and JR-CSF: 0-1500 reads. (B) Alignment of two segments from the NL4-3 and BaL genome, with a putative novel methyl receptor adenosine present in BaL shown in red. (C) Similar to panel B, except aligning two regions of NL4-3 and JR-CSF, with the two novel methyl acceptor adenosines present in JR-CSF indicated. For related data, see Figs. S1 and S4.